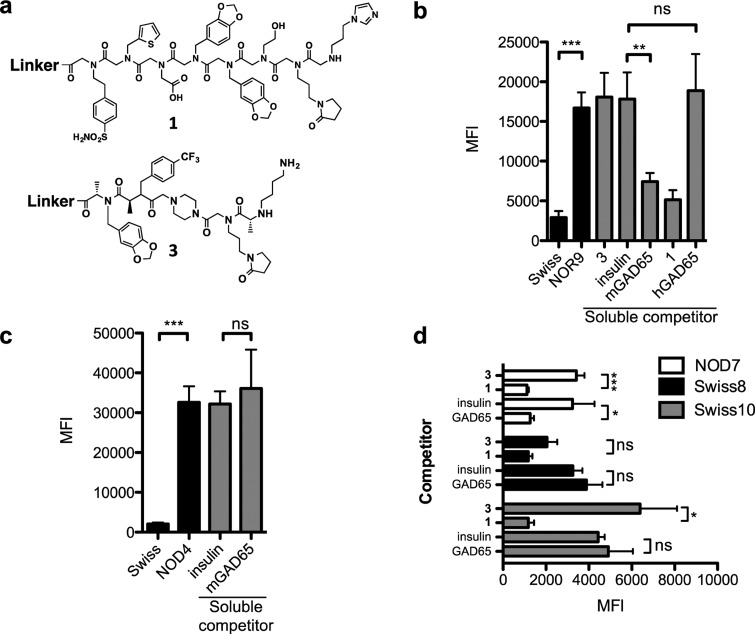
Figure 4.
Competitive binding to 1 using GAD65 as a specific competitor. (a) Chemical structures of 1 and 3. (b) Serum from pooled Swiss or NOR9 serum (500 μg mL–1) was incubated with 1 immobilized onto 10 μm TentaGel beads in a blocking buffer with no soluble competitor (black bars). Binding was monitored by flow cytometry after incubating the beads with Alexa Fluor 647-conjugated secondary antibody. Competitive binding (gray bars) to 1 was performed by preincubating 500 μg mL–1 NOR9 serum with 500 nM of either soluble 1, 3, insulin, murine GAD65 containing a GST fusion tag (mGAD65), or human GAD65 containing a GST fusion tag (hGAD65). (c) Binding to TentaGel beads containing 3 was performed by diluting pooled Swiss or NOD4 serum to 500 μg mL–1 in a blocking buffer (black bars). Competition binding to 3 (gray bars) was performed in the presence of 500 nM insulin or mGAD65. (d) Sera from two false positive samples, Swiss8 and Swiss10, and one true positive sample, NOD7, were diluted to 500 μg mL–1 and incubated in the presence of 500 nM 3, insulin, and murine GAD65 lacking a GST fusion tag (GAD65) for 5 min before incubating with immobilized 1 for 2 h. Beads were probed with Alexa Fluor 647-conjugated secondary antibody, and binding was quantified by flow cytometry. In all cases, data are reported as the mean ± s.d. from three experiments. Statistical significance was determined using an unpaired t test: *P < 0.05; **P < 0.01; ***P < 0.001; ns = not significant.