Figure 2.
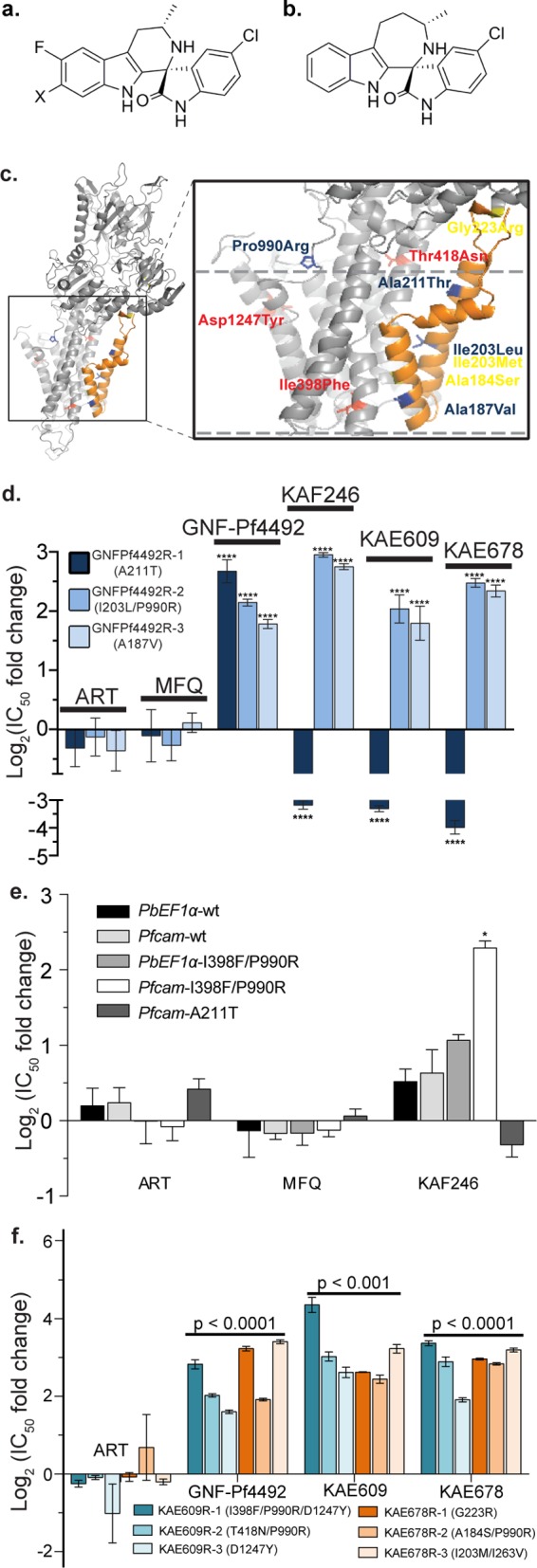
Mutations in pfatp4 confer cross-resistance between GNF-Pf4492 and the spiroindolone class. Chemical structures of representative spiroindolones (a) KAE609 when X = Cl and KAF246 when X = F. (b) KAE678 is distinguished by a 7-membered ring in the tricyclic system. (c) A PfATP4 homology model shows the location of resistance-conferring mutations specific to the aminopyrazole GNF-Pf4492 (blue) and the spiroindolones KAE609 (red) and KAE678 (yellow). Nearly all resistance-conferring mutations occur within or near PfATP4 transmembrane domains (approximated by dashed lines). Transmembranes 1 and 2 are in orange. (d) The three GNF-Pf4492-resistant lines—GNF-Pf4492R-1, GNF-Pf4492R-2, and GNF-Pf4492R-3—were tested for cross-resistance against a panel of spiroindolones (KAE609, KAE678, and KAF246). The IC50 shift is relative to the GNF-Pf4492-sensitive Dd2EF1. Artemisinin (ART) and mefloquine (MFQ) were used as controls. (e) IC50 log2 fold change in transgenic lines harboring either wild-type pfatp4 (PbEF1α-wt and Pfcam-wt) or mutated pfatp4 (PbEF1α-I398F/P990R, Pfcam-I398F/P990R or Pfcam-A211T). (f) Resistant lines (three each) were independently evolved to spiroindolone analogs KAE609 and KAE678. These lines were tested for cross-resistance against GNF-Pf4492. ART and MFQ were used as controls. Significance values were determined using one-way ANOVA followed by Dunnett’s multiple comparison post-test to test for a difference in mean log(IC50) value between each strain and the parent: ****p < 0.0001; *p < 0.01.
