FIGURE 6.
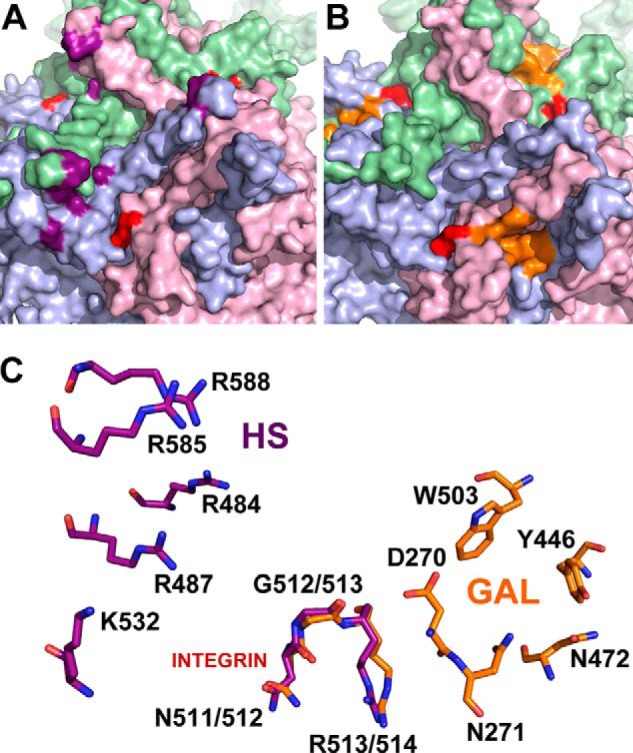
Comparison of glycan and integrin recognition motifs on AAV2 and AAV9 capsid surfaces. Exterior capsid surface representation of VP3 subunit trimers for AAV2 (A) and AAV9 (B) generated using the Oligomer Generator utility in VIPERdb-Virus Particle ExploreR2 and PyMOL. Different VP3 monomers are colored in pale green, light blue, and light pink. The surface location of heparan sulfate binding residues on AAV2 (VP1 numbering Arg484, Arg487, Lys532, Arg585, and Arg588) and the galactose binding residues on AAV9 (VP1 numbering Asp271, Asn272, Tyr446, Asn470, and Trp503) are highlighted in purple and orange, respectively. The integrin recognition motifs, 511NGR513 on AAV2 and 512NGR514 (VP1 numbering) on AAV9 are highlighted in red. C, close-up views of superposed structures of AAV2 and AAV9 trimers with one group of individual residues labeled in each. The overlapping NGR motif is structurally conserved and located in closer proximity to the AAV9 galactose footprint than the AAV2 heparan sulfate binding footprint.
