FIGURE 7.
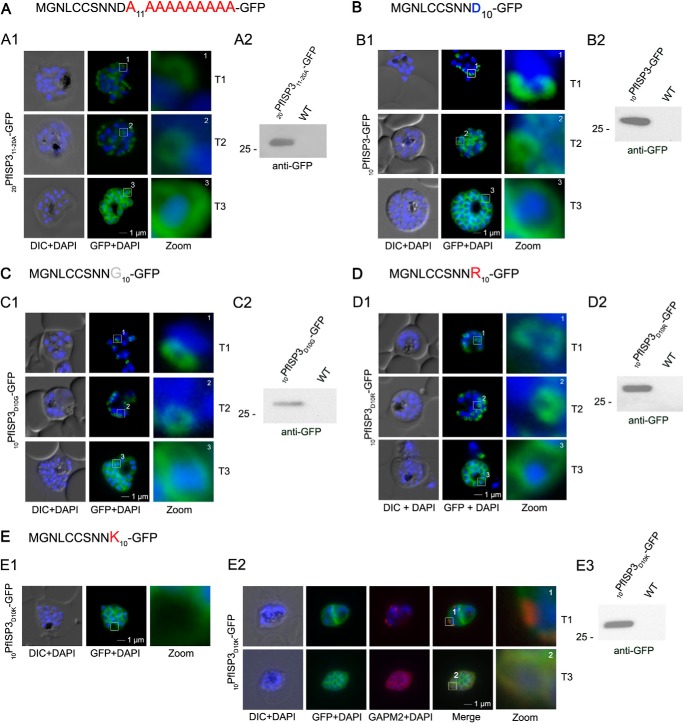
Mutational analysis of the minimal sequence requirements for IMC association of PfISP3. A and B, expression and localization of the 20-amino acid N terminus with the last 10 (20PfISP311–20A-GFP) amino acids substituted with alanines. Panel A1, 20PfISP311–20A-GFP was localized in unfixed parasites and showed an identical dynamic as the wild type protein. Panel A2, Western blot analysis of the expression using anti-GFP antibodies. B, first 10 amino acids of PfISP3 (10PfISP3-GFP) are sufficient for IMC localization (panel B1). Panel B2, Western blot analysis of the expression 10PfISP3-GFP. C and D, mutation of the sole charged residue D10 into a neutral glycine (10PfISP3D10G-GFP, panel C1) or a positively charged arginine (10PfISP3D10R-GFP, panel D1) does not interfere with IMC targeting. Western blot analysis (panels C2 and D2) of GFP fusion protein expression. E, substitution of D10 with a positively charged lysine re-directs the GFP fusion protein to the periphery of the nascent merozoites. The zoom highlights the additional association with the food vacuole membrane characteristic for plasma membrane proteins (panel E1). Co-localization was with the IMC marker GAPM2. Nascent IMC (anti-GAPM2, red) is clearly distinguishable from the peripherally associated 10PfISP3D10K-GFP in early stages (T1) and congruent in late stages (T3). Western blot analysis (panel E3) of 10PfISP3D10K-GFP. DIC, differential interference contrast.