Abstract
RNA–protein interactions are pervasive. The specificity of these interactions dictates which RNAs are controlled by what protein. Here we describe a class of revolutionary new methods that enable global views of RNA-binding specificity in vitro, for both single proteins and multiprotein complexes. In vitro methods provide insight into central issues in RNA regulation in living cells, including understanding the balance between free and bound components in cells, the basis for exclusion of binding sites in vivo, detection of binding events in the absence of discernible regulatory elements, and new approaches to targeting specific cellular RNAs by design. Comparisons of in vitro and in vivo binding provide a foundation for comprehensive understanding of the biochemistry of protein-mediated RNA regulatory networks.
Challenges and opportunities in a genome-wide view of RNA control
The biological roles of RNA–protein interactions are pervasive. They regulate mRNAs, direct assembly of RNA machines, and influence nearly every aspect of cellular function through noncoding RNAs. The immediate challenge is to determine how proteins are targeted to specific RNAs across the entire transcriptome and proteome. Understanding the connections between the entire collection of RNA-binding proteins (RBPs) and all RNAs will require analysis of their interactions in vitro and in the cell. Global analysis of RNA–protein interactions in vitro will provide a critical foundation. As ‘global’, we include both the interactions of a specific protein with a very large set of RNAs and the comprehensive analysis of all RBPs in a proteome.
Analyses of RNA–protein interactions in test tubes and cells are complementary. Indeed, the comparison of data yielded by each approach is essential to understand the biochemical basis of RNA regulatory networks, as it has been with DNA-binding proteins. In vivo methods including RNA immunoprecipitation (RIP)-ChIP, high-throughput sequencing of RNA isolated by crosslinking and immunoprecipitation (HiTS-CLIP), photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP), and individual-nucleotide resolution crosslinking and immunoprecipitation (iCLIP) have helped reveal the range and importance of RNA regulatory networks [1–4]. In each of these approaches, RNA–protein complexes are purified from cell lysates using antibodies or other affinity tags. The RNAs in the complexes are identified by either microarray or deep sequencing. In some cases (such as HiTS-CLIP, PAR-CLIP, and iCLIP), cells are UV irradiated before cell lysis to ensure that the RNA and protein are very close to one another in the cell, often (but perhaps not always) in direct contact. The tremendous power of deep sequencing enables transcriptome-wide detection of encounters between protein and RNA molecules across the entire transcriptome. These experiments require highly specific antibodies or similar-affinity reagents and provide limited quantitative information due to differences in RNA abundance and localization. An additional limitation is an inability to discriminate bound sites from functional sites. These methods can be complemented by in vitro techniques for comprehensive and quantitative assessments of binding interactions. Here we discuss recently reported methods to analyze RNA–protein interactions in vitro and consider the new challenges and opportunities they present. These frontiers include assessment of RNA–protein affinities for a large range of RNA sequences and across entire proteomes, the design of new proteins and small molecules that bind RNA with high specificity, and in-depth dissection of the biochemical basis of molecular recognition between RNAs and specific proteins in vivo.
Global approaches in vitro
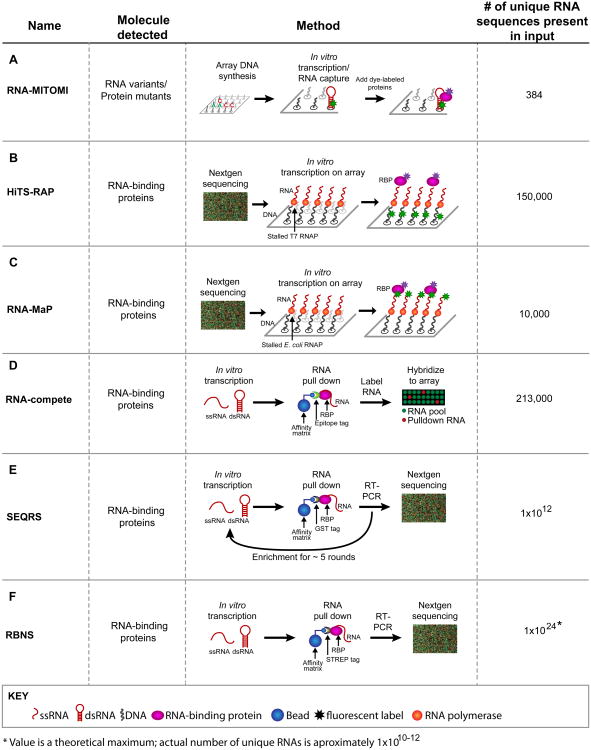
Traditionally, RNA–protein interactions have been examined using a range of quantitative approaches in vitro, electrophoretic mobility shift assays (‘gel shifts’), fluorescence anisotropy/polarization, nitrocellulose filter binding, and RNAse protection assays. These approaches, while powerful and in some cases quantitative, can analyze only a few RNA–protein interactions at a time. Recently reported methods make it possible to examine many RNA– protein interactions simultaneously in a single experiment (Figure 1) [5–11]. The exact number of RNAs analyzed varies, as does the mode of detection, which together dictate the depth to which RNA-binding preferences can be discerned (Tables 1 and 2). In vitro selection, high-throughput sequencing of RNA, and sequence specificity landscapes (SEQRS) and RNA Bind-n-Seq (RBNS), which affinity purify a protein bound to members of a large RNA library, yield information on the most diverse sets of RNA sequences. Methods that use fluorescent proteins can in principle detect protein–protein and protein–RNA interactions simultaneously and so have special strengths in dissecting the roles of protein partnerships (see next section).
Figure 1.
Methods to analyze RNA–protein and RNA–small molecule interactions. The diagrams for each method use the same color code. Red, RNA – structured (shown as a hairpin) or single stranded (squiggly lines); black, DNA; pink, RNA-binding proteins (RBPs); blue, affinity matrices; asterisks, fluorescent components. Use of arrays or deep sequencing is indicated in the diagrams. (A) In RNA mechanically induced trapping of molecular interactions (RNA-MITOMI), protein and RNA variants are analyzed by differential dye fluorescence for structure/function relationships. Both the RNA and the protein are fluorescent. (B) In high-throughput sequencing RNA affinity profiling (HiTS-RAP), RNA variants are synthesized on and tethered to an Illumina flow cell and the RBPs are detected by fluorescence at the site of DNA clusters. (C) Quantitative analysis of RNA on a massively parallel array (RNA-MaP) is similar to HiTS-RAP in that RNA variants are synthesized on a flow cell and incubated with fluorescently labeled proteins. (D) In RNA-compete, RNA–protein interactions are detected by retrieving RNAs bound to a protein of interest followed by detection of these RNAs via a microarray. Fluorescence intensity reveals binding preferences. (E) In in vitro selection, high-throughput sequencing of RNA, and sequence specificity landscapes (SEQRS), RNA–protein interactions are detected using iterative selection and high-throughput sequencing. The method is analogous to systematic evolution of ligands by exponential enrichment (SELEX) but with fewer rounds of selection and deep sequencing. (F) RNA Bind-n-Seq (RBNS) is similar to SEQRS in that RNA–protein complexes are retrieved from a complex library and detected via deep sequencing. However, unlike SEQRS only a single round of selection is used, allowing access to more RNA sequences.
Table 1. Probes, detection, and types of RNA amenable for study using new methods.
| Method | Array fabrication | Unstructured RNAa | Structured RNAa | Fluorescent probe |
|---|---|---|---|---|
| RNA-MITOMI | Yes | No | Yes | Protein and RNA |
| HiTS-RAP | No | No | Yes | Protein |
| RNA-MaP | No | No | Yes | Protein and RNA |
| RNA-compete | Yes | Yes | Yes | RNA |
| SEQRS | No | Yes | Yes | None |
| RBNS | No | Yes | No | None |
| Modified 2DCS | No | NA | NA | yes |
Demonstrated classes of RNA that have been reported.
Table 2. Demonstrated applicationsa.
| Method | Determination of unknown specificity (reported motif lengths)b | Measurement of equilibrium Kd | Measurement of off rates | Multiprotein complexesc | Genome-wide analysis |
|---|---|---|---|---|---|
| RNA-MITOMI | No | Direct | No | No | No |
| HiTS-RAP | No | Direct | No | No | No |
| RNA-MaP | No | Direct | Yes | No | No |
| RNA-compete | Yes (7) | Correlated | No | No | Yes |
| SEQRS | Yes (10) | Correlated | No | Yes | No |
| RBNS | No | Indirect | No | No | No |
| Modified 2DCS | No | No | No | NA | NA |
A comparison of the demonstrated uses of the methods, including the access they provide to the identification of previously unknown binding elements, biophysical parameters, the specificities of multiprotein complexes, and genome-wide analysis. The table includes published data only; additional developments and applications will arise.
Demonstrated capability of extracting consensus binding elements; where applicable, the maximum lengths analyzed to date are provided in parentheses.
Several of the methods provide access to multiprotein complexes through the use of multiple fluorescent proteins, although they have not been used in this way to date (see text).
RNA mechanically induced trapping of molecular interactions (RNA-MITOMI) [8]
RNA-MITOMI uses a high-throughput microfluidic platform (Figure 1A). Fluorescently labeled RNAs are immobilized on an array coated in complementary DNA oligonucleotides. Proteins labeled with a different fluorophore are incubated with the RNA pool and are used to quantify the binding of a single protein to hundreds of RNA species on the array. As currently configured, RNA-MITOMI is applicable to cases in which the RNA target is already known (e.g., histone stem-loop-binding protein).
High-throughput sequencing RNA affinity profiling (HiTS-RAP) [10] and quantitative analysis of RNA on a massively parallel array (RNA-MaP) [9]
These two methods repurpose a high-throughput instrument to measure the binding of fluorescently labeled protein (Figure 1B–C). DNA libraries encoding variants of an RNA-binding site are used as transcription templates on an Illumina sequencing flow cell. Transcription is stalled in situ, resulting in the immobilization of nascent RNAs. The association of fluorescent protein with the RNAs is then quantified. In RNA-MaP, both the RNA and the protein are fluorescently labeled, while in HiTS-RAP only the protein is fluorescent. Both methods enable quantitative measurements of biophysical parameters, including Kd, binding energy (ΔG), and off rates, for many sequences in parallel. The RNA-MaP study reports Kd and ΔG values for approximately 150 000 RNA variants of the binding site for MS2 phage coat protein, while HiTS-RAP has been used to determine the binding parameters of approximately 10 000 variants of RNA aptamers that bind two specific proteins. Kd values are obtained from millions of RNA clusters containing many copies of identical RNA molecules. RNA-MaP analysis of pairs of mutations yielded a plausible evolutionary trajectory for the evolution of an RNA–protein interaction [9].
RNA-compete [6]
A large pool of known RNA sequences is prepared from an array (Figure 1D). After incubation with a given RBP, protein-bound RNAs are fluorescently labeled and hybridized to a microarray. The motifs reported from RNA-compete are often shorter than those previously reported by multiple methods but generally contain a portion of the known consensus motifs. The approach has been used to examine the specificities of approximately 200 human RBPs and to predict binding in vivo with variable accuracy [12]. The initial RNA library comprises approximately 200 000 different RNA species with all sequences of eight or fewer nucleotides and 81% of all possible decamers. The use of microarrays rather than deep sequencing limits the dynamic range of detection.
SEQRS [7] and RBNS [13]
These approaches select RNAs that bind the protein of interest from large RNA libraries (Figure 1E,F). SEQRS is analogous to systematic evolution of ligands by exponential enrichment (SELEX) but uses only a few iterative rounds of selection so that a wide range of affinities are determined [7]. In traditional SELEX, 10–20 rounds of selection are used, leading to potentially large sequence biases [14]. In SEQRS, fewer than five rounds of selection are required and the composition of the initial starting material is used to normalize for compositional biases in the starting pool [7]. RNA libraries containing of the order of 1012 members (all possible 20-nt sequences) are incubated with immobilized protein; protein-bound RNAs are then reverse transcribed, the DNAs amplified and transcribed, and the process repeated. Their relative abundances (relative to the starting pool) are determined by deep sequencing. The method yields a wide range of RNA affinities, including low-affinity sites [15,16]. RBNS is conceptually similar but uses only a single round of selection. RBNS uses a range of protein concentrations to aid in estimating affinity. Reported SEQRS studies have yielded the preferences of natural and variant Pumilio/fem-3 mRNA-binding factor (PUF) proteins and guided the design of proteins with new specificities [17]. The reported RBNS work analyzed the affinities of three splicing factors and yielded quantitative measurements for six or seven nucleotide sequences.
Strengths and limitations
Here we consider the strengths and limitations of the methods, organized by the key challenges in the field.
Determining RNA-binding specificity
A common application of these new methods is to determine the consensus binding element of an uncharacterized RBP. A key objective here is to determine the affinities of a wide range of RNA sequences for a protein of interest. RNA-compete, SEQRS, and RBNS all accomplish this goal, but with different strengths and limitations. In SEQRS, two to seven rounds of selection yield high-quality binding models that include subsets of lower-affinity sites; by contrast, the single round of selection used in RNA-compete and RBNS yields shorter ‘core sites’ [6,7,12,13]. Similar experiments on DNA-binding proteins demonstrate that two rounds of selection yield biologically meaningful motifs for most proteins [18]. Although the core sites of many RNA-binding motifs are short (often 3–7 nt), longer sites exist and can have profound effects on binding and regulation [19–21]. Similarly, weak sites can be vital in vivo [7,22]. A priori, in analyzing a protein of unknown specificity one might want to include longer binding elements and weaker sites [7]. While the sequencing approaches sample long RNAs, after data processing, motifs yielded from each of the sequencing methods is substantially shorter, ranging from six or seven bases in RBNS and RNA-compete to ten bases in SEQRS. Deep-sequencing approaches enable assays of long RNAs containing diverse sequence permutations.
Obtaining biophysical parameters
A second critical objective lies in dissecting a known RNA–protein interaction in biochemical depth. With traditional methods, analysis of the effects of mutations in many positions on binding for either protein or RNA is painstaking and labor intensive. Parallel measurements enormously accelerate such studies. Understanding binding requires not only measurements of affinity for a wide array of RNAs, but also a quantitative analysis of biochemical constants such as Kd, ΔG, and off rate. RNA-MaP, HiTS-RAP, and RBNS provide quantitative measurements of Kd through the use of multiple protein concentrations [9,10,13]. The value of the biophysical parameters lies not only in understanding the underlying biochemistry, but also compared with measurements made in cells. The parameters provide a foundation against which in vivo can usefully be compared, as has been the case with DNA-binding proteins. For example, discrepancies can help to identify events that influence on and off rates in vivo, as can discrepancies between in vitro affinities and in vivo occupancies.
Revealing the effects of protein–protein interactions
Protein–protein interactions are likely to be critical in the assembly of larger multiprotein complexes that control RNAs (see below). Dissecting the effects of the protein partners on specificity is a major, but largely unexplored, challenge. In principle, the methods that rely on fluorescent proteins, including RNA-MITOMI, RNA-MaP, and HiTS-RAP, provide powerful ways forward, in that multiple proteins could be visualized simultaneously [8–10]. While these approaches have not yet been used for this purpose (Table 2), they are likely to be adapted to meet this need. Alternatively, deep-sequencing methods such as SEQRS have been used to identify the effects of one protein on another's specificity [7]. SEQRS does not require advance knowledge about the RNA target being analyzed but does not permit analysis of the order in which components bind or the kinetics and dynamics of their interactions.
The functions of RNA structure
Almost all of these studies are complicated by the diversity of structures that RNA can adopt. When the structures are already known, nearly all of the approaches can use RNA libraries designed to mimic and probe that structure (e.g., [8–10,23]). By contrast, when the structure of the RNA-binding site is unknown the problem is far more challenging. In random sequence libraries (as can be used in SEQRS and RBNS), the fraction of RNAs with stable higher-order structures is relatively low in the starting pool [24]. It may be possible to enrich cognate structures through iterative rounds of selection, as is well documented with conventional SELEX [15,16]. The ability to analyze long, highly structured RNAs, as can presently be approached with HiTS-RAP and RNA-MaP, will be important in dissecting the contributions of RNA structure but remains a challenging problem.
The RBPome and protein partnerships
Understanding post-transcriptional regulation on a genome-wide basis requires analysis of all trans-acting RNAs and proteins. Individual RBPs can bind hundreds to thousands of mRNAs in vivo, often with related functions. Global approaches can help reveal how these RNA–protein networks are assembled.
A recent RNA-compete study examined a large collection of RBPs including 85 human proteins [12]. This tremendous effort provided exciting insights into the diversity of RNA–protein interactions. For instance, the predictive value of biochemical affinity measurements appears to be extremely variable on a gene-by-gene basis [12]. Key challenges remain, including a deeper understanding of specificities, identification of differences among family members, characterization of regulatory RNAs [e.g., long noncoding RNAs (lncRNAs), piwi-interacting RNAs (piRNAs), miRNAs], and analysis of the remaining 600 or more human RBPs [25,26]. Large networks of RBPs collaborate and compete with one another to regulate RNA. Many proteins that bind to and regulate RNAs act in multiprotein complexes. For example, multiple proteins combine with RNAs to form functional particles, including small nuclear ribonucleoproteins (snRNPs), telomerase, and ribosomal subunits. 3′ Untranslated region (3′-UTR) regulatory complexes often involve multiple RBPs that interact with one another as well as with RNA [27].
Protein–protein partnerships can influence specificity in at least two ways. First, factors bound near one another on the RNA can promote the use of suboptimal binding sites. In Xenopus oocytes, the strength and position of binding sites for a pair of interacting proteins alters regulatory outcomes [28]. In somatic cells, combinatorial binding of AUF1 and Argonaute 2 controls the decay of selected target mRNAs in vivo [29]. Second, some interactions may result in cooperative effects on specificity, as has been observed in a PUF/cytoplasmic polyadenylation element-binding (CPEB) protein complex [7,30,31]. Similar cooperative effects may be widespread but have escaped detection for want of global methods of analysis capable of detecting subtle changes in binding preference.
In principle, RNA-MaP and HiTS-RAP are particularly well suited to identify ‘partner effects’ on binding, as multiple proteins labeled with different dyes can be examined across a wide range of RNA sequences. Thus, these approaches may provide an invaluable entry into combining the analysis of biochemical parameters such as binding constants and off rates with the effects of protein–protein interactions. These almost certainly will be important understanding the biochemical basis of RNA regulation and processing in the cell.
Excitement at the interface: in vitro meets in vivo
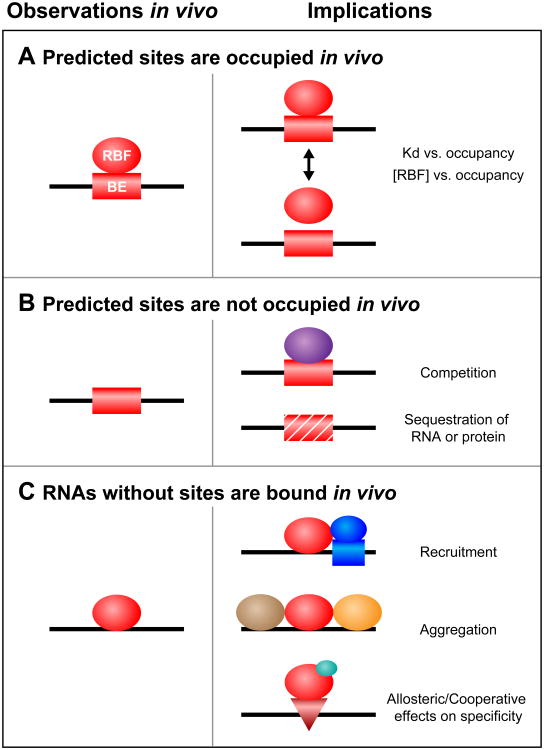
Deep sequencing of RNA following the introduction of crosslinks to associated proteins provides a way to identify RNA-binding sites (sequences, structures, or combinations thereof) in vivo [1–3]. The synergy between studies of specificity in vitro and in vivo is tremendous and is likely to be as important in understanding RNA regulation as it has been in dissecting transcriptional control [32]. At least three informative outcomes have already been documented and each category of result raises important, testable hypotheses (Figure 2).
Figure 2.
Three classes of binding element (BE) in vivo and their potential implications. (A) A BE predicted to be functional in vitro is bound in vivo by an RNA-binding factor (RBF). (B) A BE predicted to be functional in vitro is not bound in vivo, due to either competition with other factors (purple) or physical sequestration. (C) RNAs without in vitro binding sites are associated with the protein in vivo. This situation might be due to recruitment by additional factors (dark blue circle and box), aggregation (other proteins are shown in brown and orange), or the consequences of allosteric effectors (light blue circle) that drive occupancy of latent sites (red triangle).
Predicted sites are occupied in vivo (Figure 2A)
Crosslinked sites in vivo are correlated with sites identified in vitro by both RNA-compete and SEQRS approaches [7,12,33]. However, biologically meaningful binding sites are not always of the highest affinity in vitro [34]. For instance, the yeast RBP Puf5p binds two distinct motifs of different lengths and compositions [7,22]. Comparison of in vitro with in vivo immunopurification data revealed that lower-affinity modes of recognition are used in vivo [22,35].
These observations provide evidence for a range of functional binding elements beyond high-affinity sites but raise fundamental questions regarding the plasticity of binding in vivo. Over what range of affinities and protein abundance does in vitro binding directly predict in vivo occupancy? What gradations of affinity are biologically relevant? Low-affinity sites may be bound due to high concentrations of the RBP or because a protein partner that facilitates binding in vivo is absent from in vitro experiments.
From the biological perspective, the key parameter is the cellular output, such as a change in mRNA stability. It will be critical to understand how affinity and off rates relate to those outputs, particularly because function does not inevitably follow in vivo occupancy. For example, approximately 20% of the sites to which Argonaute crosslinks do not appear to mediate regulatory control in vivo [36]. Validation of bound sites by global assays of regulatory function such as RNA-seq, ribosome profiling, and proteomics will be essential to understand how occupancy relates to function [37]. Perhaps only those sites within a certain range of affinities, or within a specific sequence context, mediate regulation in the cell. If so, the use of global analysis in vitro should provide a foundation to distinguish functional sites from decoys.
Predicted sites are not occupied in vivo (Figure 2B)
Heterochromatin can mask high-affinity DNA elements from DNA-binding proteins. It is likely that mRNAs can be controlled in an analogous fashion, although through different molecular means. For example, fewer than half of the predicted sites for human PUM2 in the transcriptome appear to be occupied in vivo [2]. While it is conceivable that the absence of crosslinking is an experimental artifact, it seems likely that some potential binding sites are masked, perhaps by competing RBPs, proteins, miRNAs, or secondary structures. Sites also could be hidden through localization of mRNAs into P bodies or other granules [38]. More speculatively, RNA marks, such as N6-methylation of adenine, could modulate protein binding [39]. These modifications, some of which are known to lie in UTRs, could be harbingers of a covalent RNA code that modulates binding of regulatory proteins.
RNAs without sites are bound in vivo (Figure 2C)
Many RNAs that lack binding sites nonetheless appear to bind in vivo, as judged by crosslinking and immunopurification. Assuming that these are not experimental artifacts, their in vivo associations with an RBP could arise in three ways. First, the RBP of interest might associate with the RNA indirectly, through binding to another RBP that directly contacts the RNA. The hallmark of such events would be the presence of a completely different binding element in a subset of the in vivo targets. Alternatively, sequestration could be mediated by low-complexity regions. These protein domains, which are present in many RBPs, are implicated in the formation of amyloid-like polymers [40]. These polymers could recruit multiple RBPs that do not bind the RNA directly or without any RNA-intrinsic binding specificity. Similarly, crosslinking might arise through high concentrations of the RBP and protein in a specific subcellular location, as has been discussed elsewhere [41].
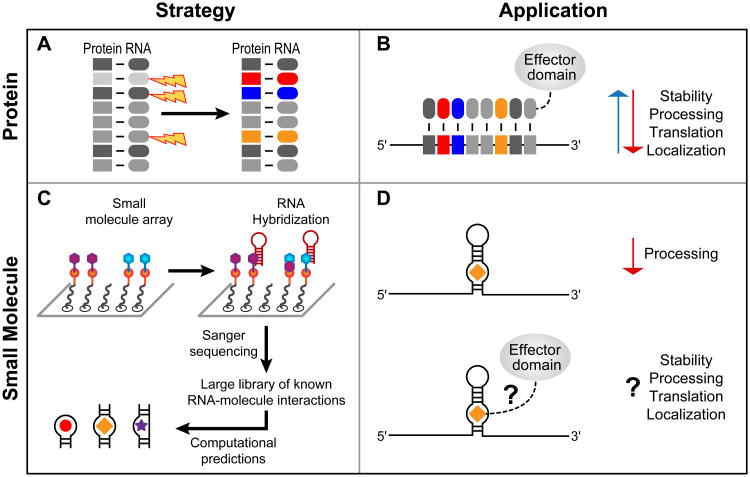
Tailored specificity: proteins and small molecules
Molecules with tailored RNA-binding preferences will enable the manipulation of RNAs for a wide variety of purposes (Figure 3A,B). For proteins, the strategy is akin to that of transcription factor-like (TAL) effectors or zinc fingers. The PUF family scaffold has proven useful for this purpose. PUF proteins bind to single-stranded RNA using short repeated α-helical modules, each of which contributes three amino acids that directly contact an RNA base. By manipulating those amino acids, new PUF proteins have been created with tailored specificities (Figure 3A). These ‘neo-PUFs’, linked to effector domains, can modify the splicing, turnover, and translation of specific mRNAs (Figure 3B) [42–48]. The specificities of natural and designed combinations of the three amino acids have been determined using a large randomized RNA library and SEQRS, yielding an ‘RNA recognition code’ [17]. This approach, which also revealed off-target effects, provides a path for the analysis of future designer proteins to control RNAs in vivo.
Figure 3.
Approaches for tailored regulation of RNA in vivo. (A) Mutagenesis of the Pumilio/fem-3 mRNA-binding factor (PUF) scaffold provides a means to target cellular RNAs [17]. Repeat units (R1–R8) specify recognition of a single RNA base and can be manipulated to change RNA targeting. (B) Fusion of PUF scaffolds to effector domains results in desired regulatory outcomes. (C) Analysis of RNA–small molecule interactions using 2D combinatorial screening (2DCS) provides a starting place for the identification of bioactive compounds. (D) Addition of chemical effector domains to compounds should enable a diversity of outcomes for the targeted RNA, as in targeted PUF design.
The importance of small molecules that bind RNA is exemplified by antibiotics that bind rRNA, metabolites that bind riboswitches, and the diverse variety of molecules that bind RNA aptamers [5,49]. SELEX has enabled the identification of RNAs that each bind specifically to their own ligand. 2D combinatorial screening (2DCS) focuses on families of ligands and may provide a route to identifying organic compounds that bind to RNAs possessing different structural motifs (Figure 3C) [11]. In one study using 2DCS, a library of structured RNAs was added to an array carrying a range of ligands and RNAs that bound specific ligands were identified by sequencing. Computational analysis of the compounds revealed the preferences of the various small molecules for structure and sequence. This enabled the prediction of compounds that proved to be effective in binding specific pre-miRNA targets, disrupting their processing (Figure 3D, top) [50]. In the future, it may be possible to attach effector domains to the ligands, as has been done with designed PUF proteins (Figure 3D, bottom). While this approach is likely to be difficult with minimally structured single-stranded sequences, it presents an exciting pathway to identifying small lead compounds that target structured RNAs.
Whatever the targeting agent, deep global analyses will be necessary to identify off-target effects, as was critical in the development of genome modification nucleases [51]. For this purpose, approaches that exploit deep sequencing are likely to be critical.
Concluding remarks
Cellular RNAs encounter a vast array of protein partners. The methods we have discussed promise to reveal how proteins find their targets, modulate one another's activity, and coordinate RNA function. The same approaches will enable targeting of specific RNAs with designer proteins and small molecules. Comprehensive analysis of RNA-binding specificity provides vital tools to explore the mechanisms of every process in which RNA molecules participate and to link the biochemistry of their interactions with their effects in the cell.
New methods probe RNA-protein interactions among whole proteomes and transcriptomes.
The RNA specificities of single proteins and protein complexes are now accessible.
The approaches yield a biochemical foundation for understanding RNA control networks.
Proteins and ligands of known specificity can be created to target specific RNAs.
Acknowledgments
The authors thank Shruti Waghray and Melanie Preston for helpful discussions and Laura Vanderploeg for help preparing the figures. M.W. is supported by National Institutes of Health (NIH) grant GM050942.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Licatalosi DD, et al. HiTS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene JD, et al. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 6.Ray D, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 7.Campbell ZT, et al. Cooperativity in RNA–protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin L, et al. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat Methods. 2012;9:1192–1194. doi: 10.1038/nmeth.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buenrostro JD, et al. Quantitative analysis of RNA–protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014;32:562–568. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tome JM, et al. Comprehensive analysis of RNA–protein interactions by high-throughput sequencing–RNA affinity profiling. Nat Methods. 2014;11:683–688. doi: 10.1038/nmeth.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran T, Disney MD. Identifying the preferred RNA motifs and chemotypes that interact by probing millions of combinations. Nat Commun. 2012;3:1125. doi: 10.1038/ncomms2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert N, et al. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho M, et al. Quantitative selection and parallel characterization of aptamers. Proc Natl Acad Sci U S A. 2013;110:18460–18465. doi: 10.1073/pnas.1315866110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriphage T4 DNA polymerase. Science. 1990;3:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 17.Campbell ZT, et al. A protein–RNA specificity code enables targeted activation of an endogenous human transcript. Nat Struct Mol Biol. 2014;21:732–738. doi: 10.1038/nsmb.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jolma A, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 20.Qiu C, et al. Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket. J Biol Chem. 2012;287:6949–6957. doi: 10.1074/jbc.M111.326264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu D, et al. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci U S A. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valley CT, et al. Patterns and plasticity in RNA–protein interactions enable recruitment of multiple proteins through a single site. Proc Natl Acad Sci U S A. 2012;109:6054–6059. doi: 10.1073/pnas.1200521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeGendre JB, et al. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem. 2013;288:2532–2545. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gevertz J, et al. In vitro RNA random pools are not structurally diverse: a computational analysis. RNA. 2005;11:853–863. doi: 10.1261/rna.7271405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Wilkie GS, et al. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 28.Pique M, et al. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, et al. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell ZT, et al. A conserved interface between PUF and CPEB proteins. J Biol Chem. 2012;287:18854–18862. doi: 10.1074/jbc.M112.352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menichelli E, et al. Biochemical characterization of the Caenorhabditis elegans FBF.CPB-1 translational regulation complex identifies conserved protein interaction hotspots. J Mol Biol. 2013;425:725–737. doi: 10.1016/j.jmb.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agius P, et al. High resolution models of transcription factor–DNA affinities improve in vitro and in vivo binding predictions. PLoS Comput Biol. 2010;6:e1000916. doi: 10.1371/journal.pcbi.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishore S, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 34.Lee MH, et al. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerber AP, et al. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi SW, et al. Argonaute HiTS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jungkamp AC, et al. In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol Cell. 2011;44:828–840. doi: 10.1016/j.molcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell SF, et al. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 40.Kato M, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riley KJ, Steitz JA. The “Observer Effect” in genome-wide surveys of protein–RNA interactions. Mol Cell. 2013;49:601–604. doi: 10.1016/j.molcel.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooke A, et al. Targeted translational regulation using the PUF protein family scaffold. Proc Natl Acad Sci U S A. 2011;108:15870–15875. doi: 10.1073/pnas.1105151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, et al. Engineering splicing factors with designed specificities. Nat Methods. 2009;6:825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu G, Hall TM. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011;19:361–367. doi: 10.1016/j.str.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 2013;280:3755–3767. doi: 10.1111/febs.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choudhury R, et al. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong S, et al. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J Biol Chem. 2011;286:26732–26742. doi: 10.1074/jbc.M111.244889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22:279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velagapudi SP, et al. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]