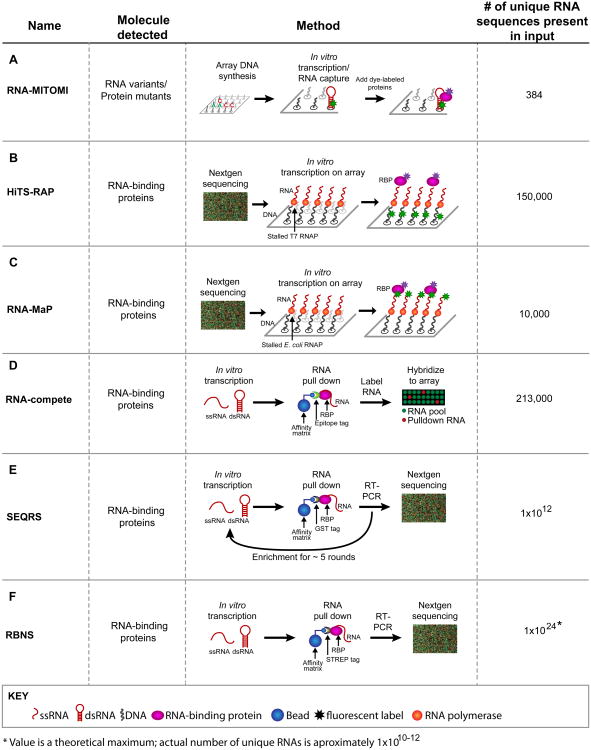
Figure 1.
Methods to analyze RNA–protein and RNA–small molecule interactions. The diagrams for each method use the same color code. Red, RNA – structured (shown as a hairpin) or single stranded (squiggly lines); black, DNA; pink, RNA-binding proteins (RBPs); blue, affinity matrices; asterisks, fluorescent components. Use of arrays or deep sequencing is indicated in the diagrams. (A) In RNA mechanically induced trapping of molecular interactions (RNA-MITOMI), protein and RNA variants are analyzed by differential dye fluorescence for structure/function relationships. Both the RNA and the protein are fluorescent. (B) In high-throughput sequencing RNA affinity profiling (HiTS-RAP), RNA variants are synthesized on and tethered to an Illumina flow cell and the RBPs are detected by fluorescence at the site of DNA clusters. (C) Quantitative analysis of RNA on a massively parallel array (RNA-MaP) is similar to HiTS-RAP in that RNA variants are synthesized on a flow cell and incubated with fluorescently labeled proteins. (D) In RNA-compete, RNA–protein interactions are detected by retrieving RNAs bound to a protein of interest followed by detection of these RNAs via a microarray. Fluorescence intensity reveals binding preferences. (E) In in vitro selection, high-throughput sequencing of RNA, and sequence specificity landscapes (SEQRS), RNA–protein interactions are detected using iterative selection and high-throughput sequencing. The method is analogous to systematic evolution of ligands by exponential enrichment (SELEX) but with fewer rounds of selection and deep sequencing. (F) RNA Bind-n-Seq (RBNS) is similar to SEQRS in that RNA–protein complexes are retrieved from a complex library and detected via deep sequencing. However, unlike SEQRS only a single round of selection is used, allowing access to more RNA sequences.