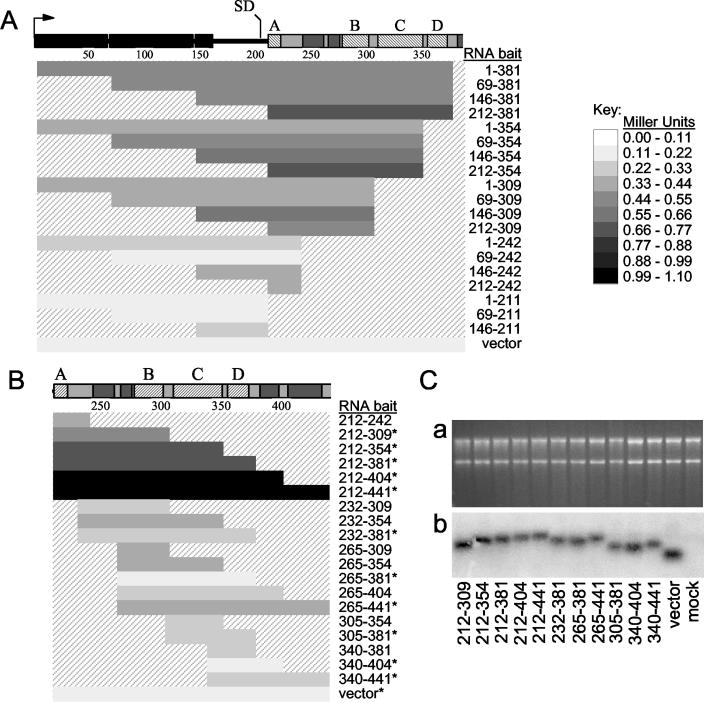
FIG. 3.
Quantitative mapping of MMLV ψ-binding activity in the yeast three-hybrid system. (A and B) Various RNA baits were tested for Gag interaction in the yeast three-hybrid system. The bars correspond to the MMLV sequences tested in the RNA baits, with nucleotide positions relative to the genome shown to the right of the bars. Reporter activation observed between the RNA bait and Gag prey in yeast, as quantified by β-galactosidase assay, is indicated by the shading of each bar. A scale relating the strength of reporter activation in Miller units to the shading of the bars is indicated to the right of the panel. Values are averages of at least three independent measurements, and standard deviations were within 10%. The RNA expression vector with no cloned MMLV sequences was tested to represent the nonspecific RNA-binding activity of Gag prey. (A) Analysis of RNA regions 5′ of the viral SD with and without Ψ sequences. (B) Mapping of Gag-interacting regions within MMLV Ψ. The RNA baits that were also quantified by Northern blot analysis (as shown in panel C) are indicated by asterisks. (C) Northern blot analysis of RNA bait expression in yeast. (a) Ethidium bromide-stained agarose gel of total yeast RNA analyzed by Northern blotting. The blot was hybridized with a radioactive probe specific for the MS2 coat protein-binding region, present in all baits, and exposed for autoradiography (b). The mock lane contains RNA from yeast cells not transformed with RNA expression vector.