Abstract
The trypomastigote small surface antigen (TSSA) is a mucin-like molecule from Trypanosoma cruzi, the etiological agent of Chagas disease, which displays amino acid polymorphisms in parasite isolates. TSSA expression is restricted to the surface of infective cell-derived trypomastigotes, where it functions as an adhesin and engages surface receptors on the host cell as a prerequisite for parasite internalization. Previous results have established TSSA-CL, the isoform encoded by the CL Brener clone, as an appealing candidate for use in serology-based diagnostics for Chagas disease. Here, we used a combination of peptide- and recombinant protein-based tools to map the antigenic structure of TSSA-CL at maximal resolution. Our results indicate the presence of different partially overlapping B-cell epitopes clustering in the central portion of TSSA-CL, which contains most of the polymorphisms found in parasite isolates. Based on these results, we assessed the serodiagnostic performance of a 21-amino-acid-long peptide that spans TSSA-CL major antigenic determinants, which was similar to the performance of the previously validated glutathione S-transferase (GST)-TSSA-CL fusion molecule. Furthermore, the tools developed for the antigenic characterization of the TSSA antigen were also used to explore other potential diagnostic applications of the anti-TSSA humoral response in Chagasic patients. Overall, our present results provide additional insights into the antigenic structure of TSSA-CL and support this molecule as an excellent target for molecular intervention in Chagas disease.
INTRODUCTION
Chagas disease is a major health and economic problem in Latin America, for which no vaccine or appropriate drugs for large-scale public health interventions are yet available (1). It is caused by the protozoan Trypanosoma cruzi, found throughout the American continents in a variety of wild and domestic mammalian reservoirs, and it is transmitted by the bite of infected blood-sucking triatomine bugs. It is estimated that 8 to 10 million people are currently infected with T. cruzi and that up to 120 million individuals living in areas that are endemic for the parasite are at risk of infection (1). Increasing travel and immigration have also brought the risk of T. cruzi infection into countries that are not endemic for the parasite (2). Several efforts have successfully been undertaken to control transmission in Latin America, with a concomitant decrease in the actual numbers of acute vector-borne infections (3). However, humans can also become infected with T. cruzi through the ingestion of tainted food and fluids, receipt of contaminated blood transfusion or organ transplantation, and from mother-to-child during pregnancy/delivery (4).
The diagnosis of Chagas disease is challenging because it is often asymptomatic in its acute phase and evolves into a chronic stage in which the disease course takes different clinical forms (1). In addition, and due to a major decline in parasitemia during the chronic phase, the detection of T. cruzi in blood samples by direct examination, hemoculture, or xenodiagnosis is difficult. Several PCR-based procedures have been reported that, though highly specific, present suboptimal sensitivity and require technological expertise and specialized expensive laboratory equipment (5). In this framework, the detection of anti-T. cruzi antibodies remains the most effective method for demonstrating direct exposure to the parasite (6). At present, the most widely used serologic methods are indirect hemagglutination assay (IHA), indirect immunofluorescence (IIF), and enzyme-linked immunosorbent assay (ELISA) using total parasite homogenates or semipurified antigenic fractions. Despite their simplicity and low cost, these tests show variations in their reproducibility and reliability that can be attributed to poor standardization of the reagents (7). In the absence of a single reference test showing ∼100% specificity and sensitivity, the current guidelines developed by the World Health Organization advise the use of two serologic tests for reaching a conclusive diagnosis. In the case of ambiguous or discordant results, a third technique should be conducted (6).
Recombinant DNA and peptide synthesis technologies allowed the production and one-step purification of large amounts of T. cruzi immunodominant antigens (8), some of which were evaluated by way of multicenter trials and are commercially available (9, 10). These antigens minimize the extent of specificity problems. As reported, serum samples from individuals with other endemic coinfections (especially Leishmania spp.) and/or afflicted by certain autoimmune disorders cross-recognize T. cruzi antigens (6, 11).
We have shown that the polypeptide backbone of a mucin-like glycoprotein displayed on the surface of infective trypomastigote forms (TSSA [trypomastigote small surface antigen]) elicits strong antibody responses in T. cruzi-infected humans and animals (12). The TSSA protein encoded by the CL Brener clone of T. cruzi (TSSA-CL) (13) showed >87% sensitivity in a seropositive chagasic panel from Argentina and Brazil, a value which increased to >98% when only parasitologically positive samples were considered (11). Most important, TSSA-CL showed a significant increase in specificity (97.4%) compared to that of the currently used serologic assays (11).
A detailed genetic characterization of the tssa locus disclosed sequence variations among parasite strains (12, 14), which correlated with the 6 major evolutionary lineages (TcI to TcVI) that were defined for the T. cruzi species (15). Interstrain polymorphisms were shown to be focused on the central region of TSSA (12, 14) and to have a major impact on its immunogenicity and antigenicity (11, 12, 16). Different attempts at using TSSA polymorphisms to design parasite lineage-specific serologic reagents as an indirect approach to allow for the typification of infecting T. cruzi strains have been undertaken (17, 18). Although these methods can be improved, they showed good concordance with genotyping techniques (17) and support the differential predominance of T. cruzi lineages causing human infections in distinct areas that are endemic for the parasite (13, 19, 20).
In addition to its serodiagnostic potential, we showed that TSSA functions in vivo as an adhesin, engaging surface receptors and inducing signaling pathways on the host cell as a prerequisite for parasite internalization (16). Interestingly, the TSSA isoforms encoded by extant parasite evolutionary lineages exhibit differential binding and Ca2+ signaling properties upon interaction with target cells (16). Overall, the contrasting antigenic and functional features of TSSA isoforms suggest that this molecule may contribute to the differential infectivity and pathogenic and epidemiological features displayed by T. cruzi evolutionary lineages (13, 21).
In the present work, we mapped the anti-TSSA-CL humoral responses in T. cruzi-infected individuals at high resolution using peptide- and recombinant protein-based approaches.
MATERIALS AND METHODS
Ethics statement.
The T. cruzi-infected human samples were obtained from the Laboratorio de Parasitología-Chagas and the Hospital de Niños Dr. Ricardo Gutierrez (Buenos Aires, Argentina). The protocol was approved by the institutional review board of the hospital. Written informed consent was obtained for each subject, and all samples were decoded and deidentified before they were provided for research purposes.
Study population.
Serum samples were collected from infected and noninfected subjects. Clotted blood was obtained by venipuncture and analyzed for T. cruzi-specific antibodies by 2 commercially available kits, an ELISA using total parasite homogenate and IHA (both from the Wiener lab, Argentina) (22). Different panels of serum samples were used in this work. The first panel was composed of 38 samples from healthy noninfected individuals rendering negative results for the 2 serologic tests mentioned above, and these were obtained from the Laboratorio de Parasitología-Chagas, Hospital de Niños Dr. Ricardo Gutierrez, or from different blood banks: Fundación Hemocentro Buenos Aires, Hospital de Enfermedades Infecciosas Dr. Francisco Javier Muñiz, Hospital Italiano de Buenos Aires (all from Buenos Aires, Argentina), and Hospital Municipal Dr. Diego E. Thompson (San Martín, Buenos Aires, Argentina). The second panel was composed of samples collected from 91 individuals (10 to 65 years old) with the chronic phase of the disease that rendered positive results for both serologic tests. The third panel was composed of 24 samples of congenitally infected or noninfected newborns (0 to 9 months old) who were diagnosed by the microhematocrit method (23). The samples from these newborns rendered also positive results for both serologic tests and, in some cases, by PCR for parasite DNA in blood (24).
Peptide chip synthesis and screening.
The overall design, production, and screening of next-generation ultrahigh-density microarray slides (25) and the data analysis will be explained in detail elsewhere (S. J. Carmona, M. Nielsen, C. Schäfer-Nielsen, J. Altcheh, J. S. Mucci, V. Balouz, V. Tekiel, A. C. Frasch, O. Campetella, C. A. Buscaglia, and F. Agüero, unpublished data). Briefly, microarray slides containing 15mer peptides, including T. cruzi-specific peptides and other controls, were incubated overnight with 1 ml of purified immunoglobulin G (IgG) diluted to 20 μg/ml in incubation buffer (0.15 M Tris-acetate [pH 8.0], 0.1% Tween 20). IgG was purified from serum samples using the Melon Gel IgG spin purification kit (Thermo Scientific), according to the manufacturer's protocol. The purity of the recovered IgG was assessed by Coomassie brilliant blue-stained 12% SDS-PAGE gels, and the concentration was estimated by a comparison against a standard curve of purified bovine γ-globulin (Bio-Rad Laboratories) (data not shown). After washing with incubation buffer, the slides were incubated for 2 h with secondary antibody (Cy3 goat anti-human IgG; Abcam) at 1 μg/ml. After a second washing step with incubation buffer, followed by a subsequent washing step with N-methylpyrrolidone and dichloromethane, the peptide array slides were air-dried, and signals were recorded with an InnoScan 900 laser scanner (Innopsys, Carbonne, France) at 1-μm resolution, with an excitation wavelength of 532 nm. Each sample corresponded to 5 different chronic Chagasic serum samples (3 μl each) that were pooled before IgG purification, and this pooled sample was assayed in duplicate. The peptide chips were sequentially assayed, first with the negative sample (pooled IgG purified from 5 healthy subjects) and then with the positive sample. With this experimental setup, 2 data sets were obtained for each experiment, one corresponding to the readout from healthy individuals (negative control) and one corresponding to the accumulated signal of the negative plus positive samples. Positive samples were then calculated by substraction (Carmona et al., unpublished data). From the whole-chip analyses, a cutoff range from 2.6 to 3.4 arbitrary units (A.U.) of fluorescence was established, as both sensitivity and specificity were optimal (i.e., all positive controls included in the array were detected, and none of the negative controls included in the array were detected) (Carmona et al., unpublished data). Accordingly, only those peptides yielding >2.6 A.U. of fluorescence in at least one screening were recorded as positive.
Recombinant proteins.
The glutathione S-transferase (GST)-fusion protein bearing the repetitive domain of T. cruzi shed acute-phase antigen (SAPA) has been described (26, 27). The GST fusion protein bearing the central region (from residues 24 to 61/62) of Sylvio X-10 TSSA (TSSA-Sy24-61) and CL Brener TSSA (TSSA-CL24-62) have also been described (11). Variants spanning partially overlapped 15mer sequences from TSSA-CL were constructed by Taq polymerase-mediated fill-in of partially complementary forward (TSSA VI Ep n Fw) and reverse (TSSA VI Ep n Rv) oligonucleotides (see Table S1 in the supplemental material) containing BamHI and EcoRI sites on their 5′ ends, respectively. The same strategy was used to generate a second set of partially overlapping 9mer sequences from TSSA-CL. In this case, however, the fill-in reactions were performed using forward (TSSA VI Ep [n + 1] Fw) and reverse (TSSA VI Ep n Rv) oligonucleotides. All of these constructs were treated with BamHI and EcoRI and cloned into pGEX-2T vector (GE Healthcare). PCR using the pGEXfw and pGEXrev oligonucleotides (see Table S1) was carried out for the initial screening of the colonies, which were subsequently confirmed using Sanger-based sequencing on an Applied Biosystems ABI3130 capillary sequencer. The supernatants of Escherichia coli strain BL21-CodonPlus (Stratagene) cultures transformed with each construct and induced for 3 h at 28°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside (Fermentas) were purified by glutathione-Sepharose chromatography (GE) (11) and dialyzed against phosphate-buffered saline (PBS). GST and the GST-fusion molecules were quantified using the Bradford reagent (Pierce), and purity was assessed using Coomassie brilliant blue-stained SDS-PAGE.
Synthetic peptides.
Custom peptides were synthesized by GenScript. The purity (>90%) and identity of the peptides were determined by the manufacturer, using reverse-phase high-performance liquid chromatography, and confirmed by ion-spray mass spectrometry methods, respectively. Some of these peptides bear an additional Cys residue, through which they were individually coupled to maleimide-activated ovalbumin (Pierce), as described previously (28, 29). The sequence and features of the synthetic peptides used in this work are shown in Table S2 in the supplemental material.
Enzyme-linked immunosorbent assay.
An ELISA was performed using flat-bottomed 96-well Nunc-Immuno plates (Nunc, Roskilde, Denmark), as described previously (30). Briefly, antigens (either GST-fusion proteins or synthetic peptides) were dissolved in carbonate buffer (pH 9.6) as a coating buffer at 10 μg/ml. Peptides coupled to ovalbumin and parasite lysates (see below) were dissolved in the same buffer at 1 μg/ml and 100 μg/ml, respectively. The plates were coated overnight at 4°C with 100 μl of the antigen solution, washed 3 times with PBS containing 0.05% Tween 20 (PBS/T), and blocked for 1 h with 4% skim milk in PBS/T at 37°C. The plates were again washed 3 times with PBS/T prior to the addition of serum samples prepared in 4% skim milk PBS/T buffer (at a 1:500 dilution). Following incubation for 1 h at 37°C and washings with PBS/T, peroxidase-conjugated goat IgG to human IgG (Sigma) diluted 1:5,000 in 4% skim milk in PBS/T was added to the plates and incubated at 37°C for 1 h. The plates were washed and incubated with 100 μl of freshly prepared citrate-phosphate buffer (pH 5.0) containing 0.2% hydrogen peroxide, followed by 50 μl of 3,3′,5,5′-tetramethylbenzidine (Sigma). The reaction was stopped with 100 μl of 2 M sulfuric acid, and the absorbance at 450 nm was read. Each sample was assayed in triplicate, unless otherwise indicated. For immunoglobulin M (IgM) determinations, the ELISA was carried out essentially as above, except that the serum samples were incubated at a 1:100 dilution with constant orbital agitation (160 rpm), followed by the addition of peroxidase-conjugated goat IgG fraction to human IgM (Sigma) diluted 1:1,000 in 4% skim milk PBS/T buffer.
Competitive ELISA.
The serum samples were diluted up to 10 μl in PBS containing 2 μg of the indicated synthetic peptide. Following 30 min of incubation at room temperature, the serum-peptide mixtures were diluted up to 1:500 in 4% skim milk PBS/T buffer, added to TSSA-CL24-62-coated plates, and processed by ELISA, as described above. Absorbance at 450 nm in the control wells in which the serum samples were incubated for 30 min with 10 μl of PBS without peptide was taken as 100% reactivity. Under these conditions, most (if not all) of the peptide-specific antibodies were depleted (not shown), and the remaining reactivity against the TSSA-CL protein should be thus ascribed to B-cell epitopes lying outside the assayed peptide.
Data analysis.
The cutoff value for each antigen was calculated using 3 negative-control serum samples assayed in parallel. The reactivity of each serum sample was considered positive for a specific antigen when the mean − 3 standard deviations (SD) was greater than the mean + 3 SD recorded for the negative sera toward the same antigen. For dot and receiver operating characteristic (ROC) analyses (31), the results were expressed as the percentage of reactivity of the mean absorbance at 450 nm of the positive reference control serum included in each assay run. The ROC analyses were then performed using the GraphPad Prism software (version 5.01 for Windows; San Diego, CA, USA). Pairwise comparisons of the area under the ROC curve (AUC) values were performed using the MedCalc Statistical software version 13.0.6 (MedCalc Software bvba, Ostend, Belgium [http://www.medcalc.org]). Multigroup comparisons were performed using analysis of variance (ANOVA), followed by Bonferroni's correction.
RESULTS
Mapping of antigenic sequences in TSSA-CL using peptide chips.
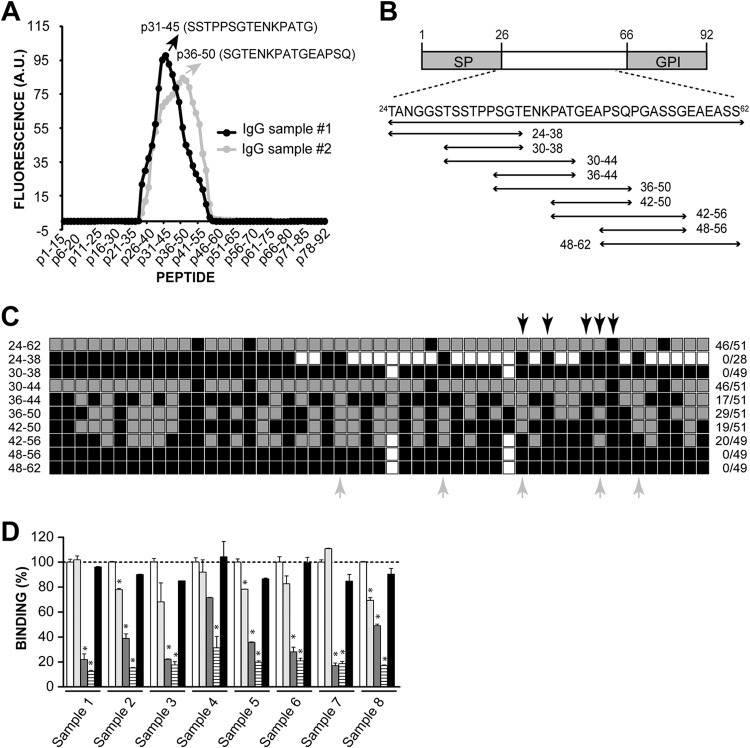
In the context of a project aimed at identifying and fine-mapping T. cruzi linear B-cell epitopes using peptide microarrays, we produced glass slides containing a tiling array of 15mer peptides derived from T. cruzi proteins (Carmona et al., unpublished data). A subset of the array comprised 78 overlapping 15mer peptides with a 14-residue overlap and 1-residue offset covering the entire TSSA-CL protein, including its predicted sorting signals (12), thus ensuring maximal resolution for mapping. It should be noted that in spite of CL Brener being a hybrid clone (13), the TSSA-CL protein deduced from both tssa genes located in tandem in an Esmeraldo-like (TcII) parental chromosome (TcCLB.507511.81 and TcCLB.507511.91) is identical. There is an additional tssa gene present in a non-Esmeraldo-like (TcIII) parental chromosome (TcCLB.508235.20) coding for a different isoform of TSSA, which was not explored in the present study. This array design was independently probed with 2 different samples, each composed of pooled IgGs purified from 5 chronic Chagasic serum samples. The fluorescence values obtained for each of the 78 TSSA-CL-derived peptides are shown in Table S3 in the supplemental material, and the recognition profile obtained by each sample is reconstructed in Fig. 1A. As shown, both samples yielded a unique and broad fluorescence peak, with positive signals for peptides p24-38 to p43-57 (sample 1) or p24-38 to p45-59 (sample 2). Together, the reactive peptides encompassed most of the mature region of TSSA-CL (i.e., the TSSA-CL sequence predicted to be displayed on the parasite surface upon processing of the N-terminal signal peptide and the C-terminal glycosylphosphatidylinositol [GPI]-anchoring motif) (Fig. 1B). Within this region, each IgG sample generated a unique profile of recognition toward individual TSSA-CL peptides, with maximal reactivity recorded either for p31-45 (sample 1) or p36-50 (sample 2) (Fig. 1A; see also Table S3). Differences in the recognition profiles can in principle be attributed to intersample, and thus interindividual, differences in the specificity of the anti-TSSA-CL humoral response (see below).
FIG 1.
Mapping of antigenic sequences in TSSA-CL. (A) Peptide chips composed of 15mer peptides overlapping by 14 residues spanning the complete sequence of TSSA-CL were probed, in duplicate, with 2 different IgG samples purified from chronic Chagasic sera. The mean reactivity toward every TSSA-CL-derived peptide (in arbitrary units [A.U.] of fluorescence) and the sequence of the peptide showing the highest reactivity for each IgG sample are indicated. (B) Schematic illustration of TSSA-CL showing the predicted signal peptide (SP) and the GPI-anchoring signal (GPI). The sequences derived from TSSA-CL that were expressed as GST-fusion molecules and the residues spanned by each construct (numbers indicate amino acid positions in each sequence relative to the initial Met) are indicated at the bottom. (C) Schematic diagram showing the anti-TSSA-CL reactivity profile obtained for each sample. Individual serum samples from chronic Chagasic patients (n = 51) were tested toward TSSA-CL deletion variants by ELISA. The gray square boxes indicate positive reactivity, and the black square boxes indicate negative reactivity. The white square boxes indicate nontested molecules. The fraction of positive serum samples/total assayed serum samples is indicated to the right. The identity of pooled sera used to purify IgG sample 1 and IgG sample 2 (see panel A) are indicated by black and gray arrows, respectively. (D) Competitive ELISAs. Eight individual serum samples from chronic Chagasic patients not tested in panel C were preincubated with 2 μg of p36-50 (light gray columns), p30-44 (dark gray columns), p30-50 (striped columns), or a scrambled version of p30-50 (black columns) for 30 min and then added to GST-TSSA24-62-coated ELISA plates. Absorbance of a control sample preincubated with PBS (white columns) was taken as 100% binding. *, significant differences between the population means (P < 0.001 after Bonferroni's correction).
Mapping of antigenic sequences in TSSA-CL using recombinant proteins.
To refine our search for antigenic sequences and to further address the variability in anti-TSSA-CL humoral responses among Chagasic patients, we undertook a complementary approach based on the use of recombinant proteins. To that end, a panel of 5 TSSA-CL deletion variants composed of partially overlapping sequences of 15 residues with a 6-amino-acid residue offset (Fig. 1B) were expressed as GST-fusion proteins in E. coli and purified through GST affinity chromatography (see Fig. S1 in the supplemental material). Together, these molecules encompassed residues 24 to 62 and hence the entire region of TSSA-CL showing reactivity in the chip assays (see Table S3 in the supplemental material). Additional GST, TSSA-CL24-62, and TSSA-Sy24-61 molecules (11) were expressed and purified in parallel to be used as controls (see Fig. S1). The serum samples collected from chronic Chagasic patients were individually assayed by ELISA against every GST-fusion molecule, and the absorbance was normalized to GST (see Fig. S2 in the supplemental material). The cutoff for each antigen was determined using 3 negative samples evaluated in parallel (not shown), and a schematic diagram showing the reactivity (positive or negative) of each Chagasic serum against every individual GST-fusion molecule is presented in Fig. 1C. As shown, TSSA-CL24-62, our positive control, was recognized by 46/51 tested samples (90.18% prevalence), whereas TSSA-Sy24-61 was not recognized by any sample (not shown), which is in close agreement with our own previous data (11, 30).
Among the 15mer variants, maximal performance was recorded for TSSA-CL30-44 (90.18%), followed by TSSA-CL36-50 (56.86%) and TSSA-CL42-56 (40.81%) (Fig. 1C). Interestingly, all serum samples that recognized TSSA-CL24-62 also reacted against TSSA-CL30-44 (Fig. 1C). Moreover, ∼80% of the reactive serum samples (36/46) showed similar antibody titers toward either molecule (i.e., Δ absorbance values, <25%; data not shown). TSSA-CL24-38 and TSSA-CL48-62, on the other hand, yielded negative results for every serum sample tested (Fig. 1C). TSSA-CL48-62 matched the sequence of p48-62 in the chip array, which was also recorded as negative for both IgG samples (see Table S3 in the supplemental material). In the case of TSSA-CL24-38, however, its matching peptide (p24-38) was recorded as positive for both IgG samples in our chip assay (see Table S3). Despite this discrepancy, which can be attributed to a higher sensitivity of the microarray, our data showed a close agreement between the two kinds of assays (peptide-chip arrays and GST-fusion protein-based ELISA).
To further analyze the anti-TSSA-CL humoral responses, we next generated a second panel of TSSA-CL deletion variants composed of 4 partially overlapping 9mer sequences with a 6-amino acid residue offset (Fig. 1B). Together, these 4 molecules encompassed the entire region (residues 30 to 56) of TSSA-CL showing reactivity by ELISA (Fig. 1C). The 9mer variants were expressed and purified, as described above (see Fig. S1 in the supplemental material), and assayed by ELISA against the same panel of chronic Chagasic serum samples (Fig. 1C; see also Fig. S2 in the supplemental material). The TSSA-CL42-50 and TSSA-CL36-44 deletion variants displayed modest prevalences (37.26% and 33.33%, respectively), whereas TSSA-CL30-38 and TSSA-CL48-56 yielded negative results for every serum sample tested. Based on these findings, we conclude that the serodiagnostic performance of the 15mer TSSA-CL30-44 relies on residues present in both 9mer variants that in concert cover TSSA-CL30-44 (TSSA-CL30-38 and TSSA-CL36-44; Fig. 1B). The same was true for the TSSA-CL36-50 molecule, which displayed a significantly increased prevalence compared with that of any of the two 9mer variants (TSSA-CL36-44 and TSSA-CL42-50) derived from it (Fig. 1B and C). The 15mer variant TSSA-CL42-56, however, showed similar reactivity toward individual serum samples and overall prevalence as those of the TSSA-CL42-50 molecule (Fig. 1C and not shown), indicating that residues 51 to 56 are largely dispensable in terms of their serodiagnosis potential. Taken together, the ELISA results revealed a differential TSSA-CL recognition signature for each serum sample (Fig. 1C), thus supporting the variability of anti-TSSA-CL humoral responses among chronic Chagasic individuals suggested by the peptide arrays assayed with pooled samples (Fig. 1A). In addition, the fact that TSSA-CL30-44 was able to recapitulate most of the diagnostic performance of TSSA-CL24-62 suggested that linear B-cell epitopes between residues 30 to 44 drive the recognition of anti-TSSA-CL antibodies elicited by chronic Chagasic patients. Additional linear B-cell epitopes between residues 36 to 50 might also contribute, though to a lesser extent, to this recognition.
To evaluate this hypothesis, we carried out competitive ELISAs. To that end, plates were coated with the recombinant TSSA-CL24-62 molecule and assayed with 8 serum samples from chronic Chagasic patients not previously evaluated and for which no a priori information regarding the TSSA-CL recognition profile was available. Before being added to the plate, the serum samples were incubated with PBS (negative control) or with different synthetic peptides derived from TSSA-CL (see Table S2 in the supplemental material). Preincubation with p36-50 had a moderate effect (up to 30% inhibition, depending on the sample), consistent with the presence of the minor linear B-cell epitope(s) underscored above (Fig. 1D). Conversely, preincubation with p30-44, which was predicted to contain the major B-cell epitope(s) of TSSA-CL (Fig. 1C; see also Fig. S2 in the supplemental material), yielded consistent and significant inhibition, which ranged from 30% to 80% (Fig. 1D). Most importantly, preincubation with a 21mer peptide that combined the sequences of both the p30-44 and p36-50 peptides (p30-50) yielded maximal inhibition (from 70 to 90%), whereas a scrambled version of this peptide (p30-50sc) used as a control did not significantly inhibit reactivity in any case (Fig. 1D). Overall, competitive ELISA studies support the hierarchical role proposed for p30-44 and p36-50 sequences in the recognition of anti-TSSA-CL antibodies and suggest that a molecule combining both sequences may be able to recapitulate the overall serodiagnostic performance (in terms of specificity and sensitivity) of TSSA-CL.
Validation of a TSSA-CL peptide as a novel tool for Chagas disease diagnosis.
To evaluate the diagnostic performance of p30-50, 2 panels of serum samples obtained from noninfected individuals (n = 38) or from patients with chronic Chagas disease (n = 70) were analyzed by ELISA. For comparison purposes, the same analysis was performed in parallel using the GST-fusion TSSA24-62 molecule (11). For both antigens, a significant difference in the overall reactivity values between the negative and positive populations was obtained (P < 0.001; Fig. 2A). Most importantly, TSSA24-62 and p30-50 displayed highly informative area under the ROC curve (AUC) values (TSSA24-62 AUC, 0.9647; p30-50 AUC, 0.9741; Fig. 2B) that yielded nonsignificant statistical differences (P = 0.4792). These results indicate that the recombinant protein TSSA24-62 and the peptide p30-50 exhibit equivalent diagnostic performance under these experimental conditions. The coupling of the p30-50 peptide to ovalbumin via its N-terminal Cys residue (see Table S2 in the supplemental material) did not affect its diagnostic performance (not shown).
FIG 2.
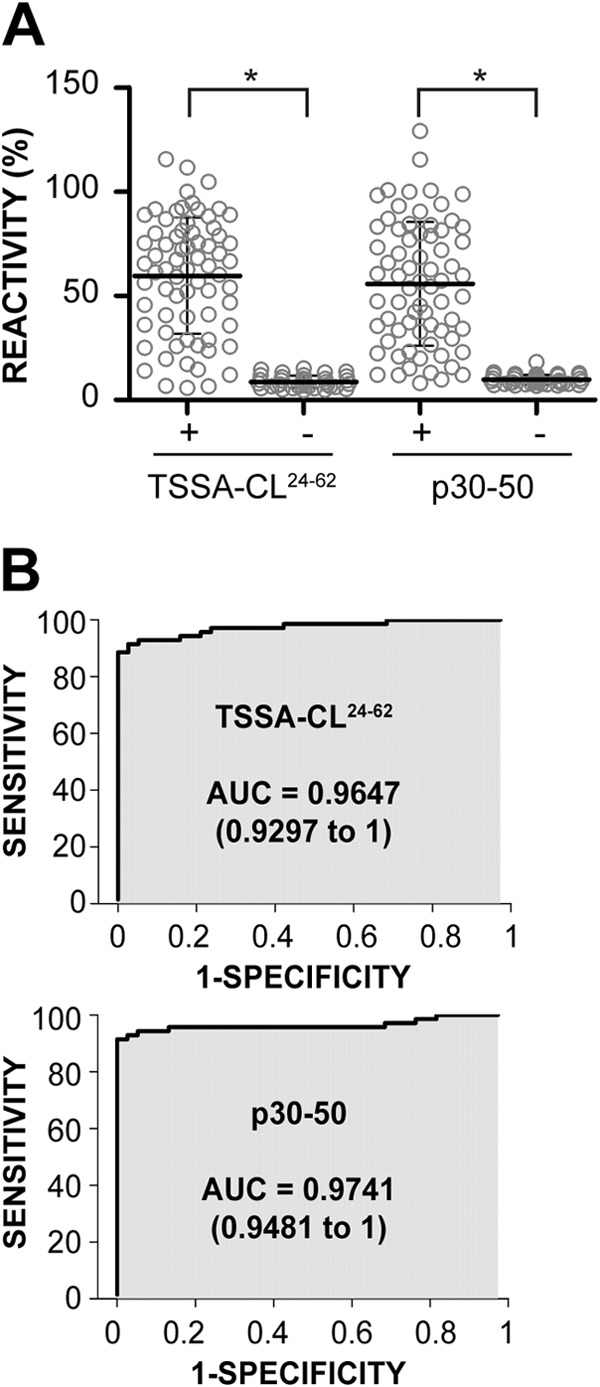
Receiver operating characteristic (ROC) curve analysis of the ELISA results using TSSA-CL-derived reagents. (A) Dot plot analysis of ELISA results using a GST-fusion protein spanning residues 24 to 62 of TSSA-CL (TSSA-CL24-62) and a synthetic peptide spanning residues 30 to 50 from the same molecule (p30-50). The ELISA plates were coated with the indicated antigen and incubated with 70 serum samples from chronic Chagas-positive individuals (+) or 38 noninfected individuals (−). The mean ± SD for each group is indicated. *, significant differences between the population means (P < 0.001 after Bonferroni's correction). (B) Reactivity values in panel A were used to generate ROC curves; the area under the ROC curve (AUC) is indicated for each antigen. The values in parentheses indicate the 95% confidence intervals.
TSSA-CL is not a suitable biomarker for diagnosing congenital infections.
The prevalence of Chagas disease in pregnant women in Latin America ranges from 5% to 40% depending on the geographical area, and the rate of vertical transmission is estimated to be 4% to 10% (32). Due to the shortage of biomarkers specific for the acute stage (32, 33), the current serologic tests are misleading in the early diagnosis of congenital T. cruzi infections, and a parasitological test should be performed on newborns for diagnosis (22). Within this framework, we evaluated the potential use of TSSA-CL in the serodiagnosis of congenital infections. Serum samples from 12 congenitally infected and 11 uninfected newborns (1 to 268 days old) born to T. cruzi-infected mothers were analyzed for IgG and IgM responses by ELISA (Table 1). When available, serum samples from their corresponding mothers were analyzed in parallel. IgM responses to TSSA-CL24-62 were only detected in 1/12 congenitally infected newborns, whereas IgM responses to a GST-fusion spanning SAPA, a T. cruzi antigen known to elicit humoral responses during the acute phase of the disease (34), were detected in 4/12 samples (Table 1). Of note, a broad IgM response to both TSSA-CL24-62 and SAPA was detected in 1 sample obtained from a Chagasic mother (Table 1).
TABLE 1.
Reactivities of TSSA-CL in pediatric samples
| Sample | Patient typea | Age (days) | Diagnosisb | IgG reactivity (A450)c |
IgM reactivity (A450)c |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GST | SAPA | TSSA | Lysate | GST | SAPA | TSSA | Lysate | ||||
| 29693 | Ch | 27 | INF | 2.808 | 0.623 | 0.815 | 0.711 | 0.394 | |||
| 29615 | M | INF | 1.694 | NDd | ND | ||||||
| 23879 | Ch | 6 | INF | 0.634 | 1.931 | 2.038 | |||||
| 25478 | M | INF | 2.270 | ND | ND | ||||||
| 24511 | Ch | 37 | INF | 1.387 | 0.522 | 0.969 | |||||
| 24510 | M | INF | 1.014 | ND | ND | ||||||
| 28036 | Ch | 32 | INF | 1.210 | 0.669 | 0.813 | |||||
| 27923 | M | INF | 0.416 | 2.113 | ND | ND | |||||
| 24520 | Ch | 6 | INF | 0.725 | 1.341 | 1.375 | |||||
| 24707 | M | INF | 0.763 | 1.640 | ND | ND | |||||
| 23078 | Ch | 12 | INF | 0.914 | 2.303 | 2.194 | |||||
| 23077 | M | INF | 0.491 | 2.616 | ND | ND | |||||
| 31511 | Ch | 8 | NI | 0.746 | 2.126 | 2.237 | |||||
| 30471 | Ch | 268 | NI | ||||||||
| 32020e | M | INF | 0.926 | 2.069 | ND | 0.315 | 0.727 | ND | |||
| 32931 | Ch | 24 | NI | 0.259 | 1.238 | ||||||
| 32929 | M | INF | 0.594 | ND | ND | ||||||
| 24467 | Ch | 24 | NI | 0.403 | |||||||
| 24468 | M | INF | 0.491 | 2.616 | ND | ND | |||||
| 30681 | Ch | 21 | NI | 1.240 | |||||||
| 30682 | M | INF | 0.570 | ND | ND | ||||||
| 33263 | Ch | 84 | NI | ||||||||
| 33267 | M | INF | 1.025 | ND | ND | ||||||
| 33341 | Ch | 233 | NI | ||||||||
| 32008 | M | INF | ND | ND | |||||||
| 34829 | Ch | 64 | NI | ||||||||
| 34828 | M | NI | ND | ND | |||||||
| 23176 | Ch | 17 | INF | 1.045 | 0.415 | 0.337 | |||||
| 25093 | Ch | 15 | INF | 0.879 | 0.657 | 1.091 | 0.280 | ||||
| 28992 | Ch | 129 | INF | 2.169 | 0.486 | 0.846 | |||||
| 33798 | Ch | 1 | INF | 0.909 | 0.764 | ||||||
| 25513 | Ch | 6 | INF | ||||||||
| 31511 | Ch | 6 | INF | 0.327 | 0.547 | 0.543 | 0.504 | ||||
| 34123 | Ch | 2 | NI | 1.957 | 0.775 | ||||||
| 34892 | Ch | 256 | NI | ||||||||
| 34226 | Ch | 239 | NI | ||||||||
| 30974 | Ch | 43 | NI | ||||||||
Ch, child; M, corresponding mother.
Patients were diagnosed as described under Materials and Methods and classified accordingly as infected (INF) or noninfected (NI).
The results for the reactive samples are expressed as the means from 3 independent experiments performed in duplicate (SD values were not >10% of the means in any case [not shown]).
ND, not determined.
Patient 32020 is the mother for both 31511 and 30471.
When analyzed for IgG antibodies, 11/12 Chagasic mothers had detectable responses toward TSSA-CL24-62, whereas 5 of them also showed reactivity to SAPA (Table 1). These values are compatible with the prevalence of either antigen in patients in the chronic stage of the disease (11, 30, 34). An analysis of the serum samples from the corresponding newborns showed IgG responses against total parasite lysates (10/12) and against TSSA-CL24-62 (8/12) and SAPA (7/12) (Table 1). It is noteworthy that the IgG titers against SAPA were consistently higher in the infected newborns than those in the chronically infected mothers (Table 1), which cannot be easily explained by transplacentally transferred antibodies from their mothers. Moreover, 3 of the infected newborns displaying IgG responses against SAPA were born to SAPA-nonrespondent mothers, which is a clear indication of the active production of antibodies due to infection (Table 1) (34). Conversely, anti-TSSA-CL24-62 IgG titers were consistently higher in chronically infected mothers than those in their infected or uninfected newborns (Table 1). A similar analysis using serial dilutions of selected paired serum samples were performed to further address this issue (see Fig. S3 in the supplemental material). Together, our data indicate that TSSA-CL is not an acute-phase antigen in Chagas disease and hence does not provide a suitable biomarker for the serologic diagnosis of congenital infections.
DISCUSSION
The lack of highly accurate methods to diagnose Chagas disease hampers the correct identification and treatment of T. cruzi-infected individuals and restricts the evaluation of the effectiveness of initiatives aimed at developing novel chemotherapy or vaccination strategies (6). To overcome this limitation, different approaches intended for the identification of T. cruzi immunodominant molecules have been undertaken (35–39). However, B-cell epitope mapping by high-resolution scanning of antibody-binding specificities toward these antigens, which is critical in guiding the design and production of customized serodiagnostic reagents with improved specificity (40), are still scarce.
To gain further insights into the antigenic structure of TSSA-CL, we undertook in this study an exhaustive mapping of immunoreactive sequences using a combination of peptide- and recombinant protein-based approaches. Overall, our results indicate a high variability in the anti-TSSA-CL humoral responses among chronic Chagasic patients, which translates into differential antibody recognition signatures (Fig. 1C). This is compatible with the existence of a broad antigenic region in TSSA-CL, spanning most of its mature region, which is likely composed of distinct and partially overlapping linear B-cell epitopes. Despite this interindividual variability, our data indicate that a linear B-cell epitope(s) between residues 30 to 44 and, to a lesser extent, between residues 36 to 50, drives the recognition of anti-TSSA-CL antibodies elicited by chronic Chagasic patients. Indeed, a synthetic peptide combining both sequences (p30-50) is able to recapitulate the overall serodiagnostic performance of TSSA-CL24-62 observed in previous studies (11). Peptide p30-50 may provide for the development of reproducible, easily manufactured, and standardized reagents for rapid, high-throughput, and low-cost serodiagnosis of Chagas disease. Additional advantages over a conventional GST-fusion antigen can be envisaged. On one hand, p30-50 may provide an advantage in terms of the specificity of the diagnostic assay. As previously shown (11), when used under more stringent conditions, a minimal fraction of Chagas-negative serum samples from patients affected by autoimmune and/or other endemic coinfection diseases reacted against bacterium-derived contaminants and/or the GST motif in TSSA-CL24-62. On the other hand, p30-50 may contribute to improve the robustness of TSSA-based serologic methods in molecular epidemiology studies (17, 18). The use of a more defined reagent lacking most of the sequences conserved across TSSA isoforms (14) may diminish its level of cross-recognition by antibodies elicited toward a different TSSA isoform.
The interindividual variability in anti-TSSA-CL humoral responses disclosed here might be further explored in other areas of Chagas disease research (6, 41). Although our data indicate that TSSA does not provide a suitable biomarker for the serologic diagnosis of congenital infections, a comparative evaluation of more defined populations of chronic Chagasic individuals (i.e., those with different clinical forms, times postinfection, or times upon undergoing chemotherapy) might lead to the identification of differential antibody recognition signatures.
In summary, our present results provide additional insights into TSSA-CL antigenic structure. Our findings, together with the identification of novel T. cruzi antigens as a result of ongoing screenings of peptide microarrays (42, 43; Carmona et al., unpublished data), will contribute to the definition of a highly effective antigenic panel for Chagas disease serodiagnosis.
ACKNOWLEDGMENTS
We thank the Fundación Hemocentro Buenos Aires (Buenos Aires, Argentina), Hospital de Enfermedades Infecciosas Dr. Francisco Javier Muñiz (Buenos Aires, Argentina), Hospital Italiano de Buenos Aires (Buenos Aires, Argentina), and Hospital Municipal Dr. Diego E. Thompson (San Martín, Buenos Aires, Argentina) for providing the serum samples. We also thank Liliana Sferco for culturing parasites, Luciano Melli and Karina Pasquevich for assistance with protein purification and statistical methods, Gabriela Figini for IgG purification, and Valeria Tekiel for trypomastigote lysates. We are also indebted to Andres Ciocchini (IIB-INTECh) for critical reading of the manuscript.
This investigation received financial support from the ANPCyT (to F.A., J.A., and C.A.B.) and the Fundación Bunge y Born (to C.A.B.).
V.B. holds a fellowship from the Consejo Inter-Universitario Nacional (Argentina). R.V. and N.G. hold fellowships from the ANPCyT and Ministry of Health (Argentina), respectively. M.d.l.M.C. and S.J.C. hold fellowships from the Argentinean Research Council (CONICET), and F.A. and C.A.B. are career investigators from the same institution. J.A. is a clinical investigator from the Hospital de Niños Dr. Ricardo Gutiérrez (Ministerio de Salud, Gobierno de la Ciudad de Buenos Aires).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00684-14.
REFERENCES
- 1.Rassi A Jr, Rassi A, Marin-Neto JA. 2010. Chagas disease. Lancet 375:1388–1402. doi: 10.1016/S0140-6736(10)60061-X. [DOI] [PubMed] [Google Scholar]
- 2.Gascon J, Bern C, Pinazo MJ. 2010. Chagas disease in Spain, the United States and other non-endemic countries. Acta Trop 115:22–27. doi: 10.1016/j.actatropica.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Reithinger R, Tarleton RL, Urbina JA, Kitron U, Gurtler RE. 2009. Eliminating Chagas disease: challenges and a roadmap. BMJ 338:b1283. doi: 10.1136/bmj.b1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muñoz-Calderón A, Diaz-Bello Z, Valladares B, Noya O, Lopez MC, Alarcon de Noya B, Thomas MC. 2013. Oral transmission of Chagas disease: typing of Trypanosoma cruzi from five outbreaks occurred in Venezuela shows multiclonal and common infections in patients, vectors and reservoirs. Infect Genet Evol 17:113–122. doi: 10.1016/j.meegid.2013.03.036. [DOI] [PubMed] [Google Scholar]
- 5.Brasil PEAA, De Castro L, Hasslocher-Moreno AM, Sangenis LH, Braga JU. 2010. ELISA versus PCR for diagnosis of chronic Chagas disease: systematic review and meta-analysis. BMC Infect Dis 10:337. doi: 10.1186/1471-2334-10-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomes YM, Lorena VM, Luquetti AO. 2009. Diagnosis of Chagas disease: what has been achieved? What remains to be done with regard to diagnosis and follow up studies? Mem Inst Oswaldo Cruz 104(Suppl 1):S115–S121. doi: 10.1590/S0074-02762009000900017. [DOI] [PubMed] [Google Scholar]
- 7.Afonso AM, Ebell MH, Tarleton RL. 2012. A systematic review of high quality diagnostic tests for Chagas disease. PLoS Negl Trop Dis 6:e1881. doi: 10.1371/journal.pntd.0001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silveira JF, Umezawa ES, Luquetti AO. 2001. Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol 17:286–291. doi: 10.1016/S1471-4922(01)01897-9. [DOI] [PubMed] [Google Scholar]
- 9.Caballero ZC, Sousa OE, Marques WP, Saez-Alquezar A, Umezawa ES. 2007. Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin Vaccine Immunol 14:1045–1049. doi: 10.1128/CVI.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reithinger R, Grijalva MJ, Chiriboga RF, de Noya BA, Torres JR, Pavia-Ruz N, Manrique-Saide P, Cardinal MV, Gurtler RE. 2010. Rapid detection of Trypanosoma cruzi in human serum by use of an immunochromatographic dipstick test. J Clin Microbiol 48:3003–3007. doi: 10.1128/JCM.02474-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Marchi CR, Di Noia JM, Frasch AC, Amato Neto V, Almeida IC, Buscaglia CA. 2011. Evaluation of a recombinant Trypanosoma cruzi mucin-like antigen for serodiagnosis of Chagas' disease. Clin Vaccine Immunol 18:1850–1855. doi: 10.1128/CVI.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Noia JM, Buscaglia CA, De Marchi CR, Almeida IC, Frasch AC. 2002. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas' disease is due to a single parasite lineage. J Exp Med 195:401–413. doi: 10.1084/jem.20011433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR. 2012. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12:240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya T, Brooks J, Yeo M, Carrasco HJ, Lewis MD, Llewellyn MS, Miles MA. 2010. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int J Parasitol 40:921–928. doi: 10.1016/j.ijpara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG, Second Satellite Meeting . 2009. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz 104:1051–1054. doi: 10.1590/S0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 16.Cánepa GE, Degese MS, Budu A, Garcia CR, Buscaglia CA. 2012. Involvement of TSSA (trypomastigote small surface antigen) in Trypanosoma cruzi invasion of mammalian cells. Biochem J 444:211–218. doi: 10.1042/BJ20120074. [DOI] [PubMed] [Google Scholar]
- 17.Risso MG, Sartor PA, Burgos JM, Briceno L, Rodriguez EM, Guhl F, Chavez OT, Espinoza B, Monteon VM, Russomando G, Schijman AG, Bottasso OA, Leguizamon MS. 2011. Immunological identification of Trypanosoma cruzi lineages in human infection along the endemic area. Am J Trop Med Hyg 84:78–84. doi: 10.4269/ajtmh.2011.10-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya T, Falconar AK, Luquetti AO, Costales JA, Grijalva MJ, Lewis MD, Messenger LA, Tran TT, Ramirez JD, Guhl F, Carrasco HJ, Diosque P, Garcia L, Litvinov SV, Miles MA. 2014. Development of peptide-based lineage-specific serology for chronic Chagas disease: geographical and clinical distribution of epitope recognition. PLoS Negl Trop Dis 8:e2892. doi: 10.1371/journal.pntd.0002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas JM, Lages-Silva E, Crema E, Pena SD, Macedo AM. 2005. Real time PCR strategy for the identification of major lineages of Trypanosoma cruzi directly in chronically infected human tissues. Int J Parasitol 35:411–417. doi: 10.1016/j.ijpara.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Cura CI, Lucero RH, Bisio M, Oshiro E, Formichelli LB, Burgos JM, Lejona S, Bruses BL, Hernandez DO, Severini GV, Velazquez E, Duffy T, Anchart E, Lattes R, Altcheh J, Freilij H, Diez M, Nagel C, Vigliano C, Favaloro L, Favaloro RR, Merino DE, Sosa-Estani S, Schijman AG. 2012. Trypanosoma cruzi discrete typing units in Chagas disease patients from endemic and non-endemic regions of Argentina. Parasitology 139:516–521. doi: 10.1017/S0031182011002186. [DOI] [PubMed] [Google Scholar]
- 21.Buscaglia CA, Di Noia JM. 2003. Trypanosoma cruzi clonal diversity and the epidemiology of Chagas' disease. Microbes Infect 5:419–427. doi: 10.1016/S1286-4579(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 22.Altcheh J, Moscatelli G, Moroni S, Garcia-Bournissen F, Freilij H. 2011. Adverse events after the use of benznidazole in infants and children with Chagas disease. Pediatrics 127:e212–218. doi: 10.1542/peds.2010-1172. [DOI] [PubMed] [Google Scholar]
- 23.Freilij H, Altcheh J. 1995. Congenital Chagas' disease: diagnostic and clinical aspects. Clin Infect Dis 21:551–555. doi: 10.1093/clinids/21.3.551. [DOI] [PubMed] [Google Scholar]
- 24.Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, Cura C, Auter F, Veron V, Qvarnstrom Y, Deborggraeve S, Hijar G, Zulantay I, Lucero RH, Velazquez E, Tellez T, Sanchez Leon Z, Galvao L, Nolder D, Monje Rumi M, Levi JE, Ramirez JD, Zorrilla P, Flores M, Jercic MI, Crisante G, Anez N, De Castro AM, Gonzalez CI, Acosta Viana K, Yachelini P, Torrico F, Robello C, Diosque P, Triana Chavez O, Aznar C, Russomando G, Buscher P, Assal A, Guhl F, Sosa Estani S, DaSilva A, Britto C, Luquetti A, Ladzins J. 2011. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis 5:e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buus S, Rockberg J, Forsstrom B, Nilsson P, Uhlen M, Schafer-Nielsen C. 2012. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol Cell Proteomics 11:1790–1800. doi: 10.1074/mcp.M112.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buscaglia CA, Alfonso J, Campetella O, Frasch AC. 1999. Tandem amino acid repeats from Trypanosoma cruzi shed antigens increase the half-life of proteins in blood. Blood 93:2025–2032. [PubMed] [Google Scholar]
- 27.Alvarez P, Leguizamon MS, Buscaglia CA, Pitcovsky TA, Campetella O. 2001. Multiple overlapping epitopes in the repetitive unit of the shed acute-phase antigen from Trypanosoma cruzi enhance its immunogenic properties. Infect Immun 69:7946–7949. doi: 10.1128/IAI.69.12.7946-7949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitcovsky TA, Buscaglia CA, Mucci J, Campetella O. 2002. A functional network of intramolecular cross-reacting epitopes delays the elicitation of neutralizing antibodies to Trypanosoma cruzi trans-sialidase. J Infect Dis 186:397–404. doi: 10.1086/341463. [DOI] [PubMed] [Google Scholar]
- 29.Urban I, Santurio LB, Chidichimo A, Yu H, Chen X, Mucci J, Aguero F, Buscaglia CA. 2011. Molecular diversity of the Trypanosoma cruzi TcSMUG family of mucin genes and proteins. Biochem J 438:303–313. doi: 10.1042/BJ20110683. [DOI] [PubMed] [Google Scholar]
- 30.Longhi SA, Atienza A, Perez Prados G, Buying A, Balouz V, Buscaglia CA, Santos R, Tasso LM, Bonato R, Chiale P, Pinilla C, Judkowski VA, Gomez KA. 2014. Cytokine production but lack of proliferation in peripheral blood mononuclear cells from chronic Chagas' disease cardiomyopathy patients in response to T. cruzi ribosomal P proteins. PLoS Negl Trop Dis 8:e2906. doi: 10.1371/journal.pntd.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greiner M, Pfeiffer D, Smith RD. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med 45:23–41. doi: 10.1016/S0167-5877(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 32.Russomando G, Sanchez Z, Meza G, de Guillen Y. 2010. Shed acute-phase antigen protein in an ELISA system for unequivocal diagnosis of congenital Chagas disease. Expert Rev Mol Diagn 10:705–707. doi: 10.1586/erm.10.70. [DOI] [PubMed] [Google Scholar]
- 33.Affranchino JL, Ibanez CF, Luquetti AO, Rassi A, Reyes MB, Macina RA, Aslund L, Pettersson U, Frasch AC. 1989. Identification of a Trypanosoma cruzi antigen that is shed during the acute phase of Chagas' disease. Mol Biochem Parasitol 34:221–228. doi: 10.1016/0166-6851(89)90050-9. [DOI] [PubMed] [Google Scholar]
- 34.Reyes MB, Lorca M, Munoz P, Frasch AC. 1990. Fetal IgG specificities against Trypanosoma cruzi antigens in infected newborns. Proc Natl Acad Sci U S A 87:2846–2850. doi: 10.1073/pnas.87.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ibañez CF, Affranchino JL, Frasch AC. 1987. Antigenic determinants of Trypanosoma cruzi defined by cloning of parasite DNA. Mol Biochem Parasitol 25:175–184. doi: 10.1016/0166-6851(87)90006-5. [DOI] [PubMed] [Google Scholar]
- 36.Burns JM Jr, Shreffler WG, Rosman DE, Sleath PR, March CJ, Reed SG. 1992. Identification and synthesis of a major conserved antigenic epitope of Trypanosoma cruzi. Proc Natl Acad Sci U S A 89:1239–1243. doi: 10.1073/pnas.89.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia V, Sinha M, Luxon B, Garg N. 2004. Utility of the Trypanosoma cruzi sequence database for identification of potential vaccine candidates by in silico and in vitro screening. Infect Immun 72:6245–6254. doi: 10.1128/IAI.72.11.6245-6254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goto Y, Carter D, Reed SG. 2008. Immunological dominance of Trypanosoma cruzi tandem repeat proteins. Infect Immun 76:3967–3974. doi: 10.1128/IAI.00604-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooley G, Etheridge RD, Boehlke C, Bundy B, Weatherly DB, Minning T, Haney M, Postan M, Laucella S, Tarleton RL. 2008. High throughput selection of effective serodiagnostics for Trypanosoma cruzi infection. PLoS Negl Trop Dis 2:e316. doi: 10.1371/journal.pntd.0000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flower DR. 2013. Designing immunogenic peptides. Nat Chem Biol 9:749–753. doi: 10.1038/nchembio.1383. [DOI] [PubMed] [Google Scholar]
- 41.Pinazo MJ, Thomas MC, Bua J, Perrone A, Schijman AG, Viotti RJ, Ramsey JM, Ribeiro I, Sosa-Estani S, Lopez MC, Gascon J. 2014. Biological markers for evaluating therapeutic efficacy in Chagas disease, a systematic review. Expert Rev Anti Infect Ther 12:479–496. doi: 10.1586/14787210.2014.899150. [DOI] [PubMed] [Google Scholar]
- 42.Mendes TA, Reis Cunha JL, de Almeida Lourdes R, Rodrigues Luiz GF, Lemos LD, dos Santos AR, da Camara AC, Galvao LM, Bern C, Gilman RH, Fujiwara RT, Gazzinelli RT, Bartholomeu DC. 2013. Identification of strain-specific B-cell epitopes in Trypanosoma cruzi using genome-scale epitope prediction and high-throughput immunoscreening with peptide arrays. PLoS Negl Trop Dis 7:e2524. doi: 10.1371/journal.pntd.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reis-Cunha JL, de Oliveira Mendes TA, de Almeida Lourdes R, dos Santos Ribeiro DR, Machado-de-Avila RA, de Oliveira Tavares M, Lemos DS, Camara AC, Olortegui CC, de Lana M, da Cunha Galvao LM, Fujiwara RT, Bartholomeu DC. 2014. Genome-wide screening and identification of new Trypanosoma cruzi antigens with potential application for chronic Chagas disease diagnosis. PLoS One 9:e106304. doi: 10.1371/journal.pone.0106304. [DOI] [PMC free article] [PubMed] [Google Scholar]