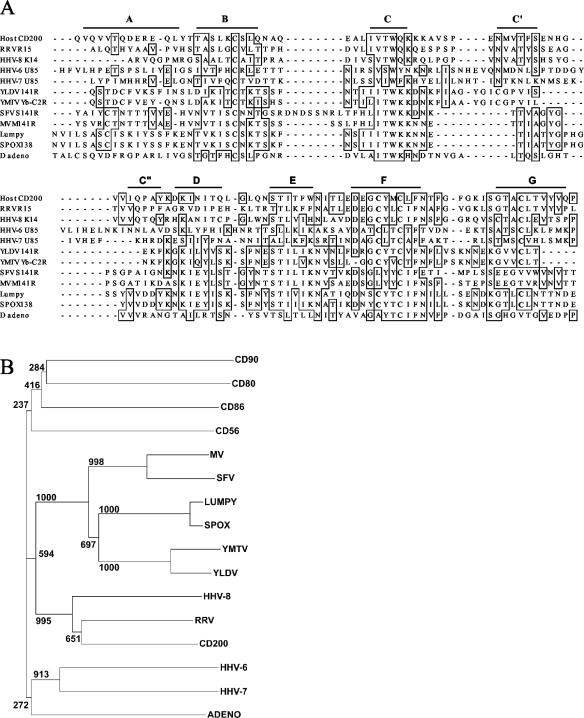
FIG. 1.
Alignment and phylogenetic analysis of CD200 and viral CD200-like ORFs. (A) Alignment of the predicted Ig V-like domains of CD200 and viral CD200 homologues. The sequences shown (with GenBank [unless stated] accession numbers) are as follows: human CD200 (Swissprot P41217), R15 from rhesus macaque rhadinovirus (RRV) (AF083501), K14 from HHV-8 (U75698), U85 from HHV-6 (X83413), U85 from HHV-7 (U43400), 141R from Yaba-like disease virus (YLDV) (NP_073526), Yb-C2R from Yaba monkey tumor virus (YMTV) (AB025319), LSDV138 from lumpy skin disease virus (Lumpy) (NP_150572), S141R from Shope fibroma virus (SFV) (AF170722), M141R from myxomavirus (MV) strain Lausanne (AF170726); sheep pox virus (SPOX) ORF138 (NP_659708), and duck adenovirus (D adeno) ORF4 (DAA00544). The bars predict the extent of the beta-strands characteristic of the Ig fold by comparison to solved structures. The cysteines that are thought to form an unusual F-to-G strand disulfide are in boldface, and residues identical in four or more sequences are boxed. (B) The N-terminal IgSF domains of the viral sequences in panel A, indicated by the virus name, were aligned to the N-terminal domains of human CD56, CD80, CD90, and CD200 by using ClustalW (38) before manual refinement and then were trimmed to remove the widely variant A strands. A neighbor-joining tree was constructed by using 1,000 bootstrap trials.