Fig. 1.
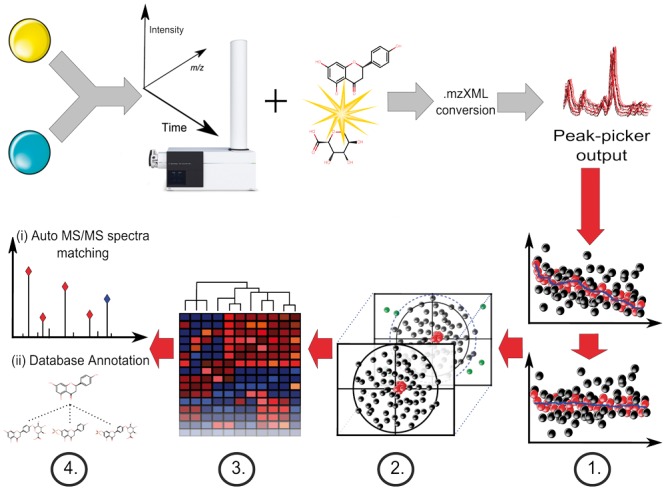
Data acquisition and MetMSLine data processing workflow (Steps 1–4). Sample preparation (e.g. urine dilution) is followed by untargeted MS and MS/MS data acquisition in sequence and peak picking softwares MetMSLine then performs sequentially: signal drift correction and pre-processing (Step 1), automatic PCA-based outlier removal (Step 2) (samples = black, QCs = red, outliers = green), automatic iterative regression based on continuous Y-variables supplied and cluster ion identification (Step 3) and final identification by data-dependent MS/MS and database matching (Step 4)
