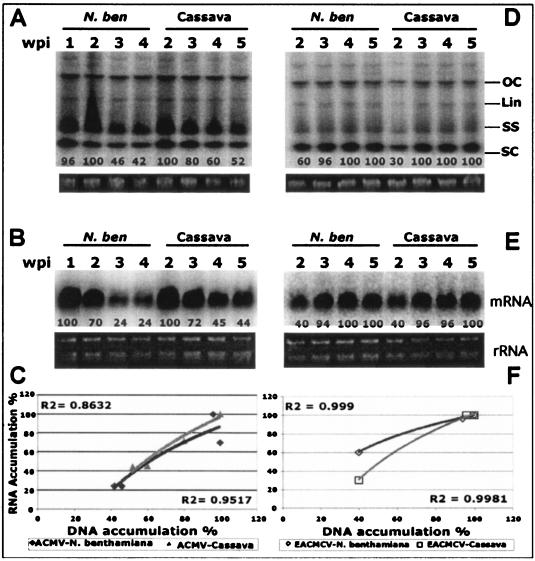
FIG. 2.
Levels of viral DNA and mRNA accumulation in ACMV-[CM] and EACMCV-infected N. benthamiana and cassava. (A and D) Southern blot analysis ACMV-[CM] DNA accumulation at 1, 2, 3, and 4 wpi in N. benthamiana and at 2, 3, 4, and 5 wpi in cassava (A) and EACMCV DNA accumulation at 2, 3, 4, and 5 wpi in N. benthamiana and cassava (D). Each lane was loaded with 4 μg of total genomic DNA isolated from virus-infected plants. Different viral DNA forms—supercoiled (SC), single stranded (SS), linear (Lin), and open circular (OC)—are indicated. Ethidium bromide-stained gels at the bottom of each blot serve as loading control. (B and E) Northern blot analysis of ACMV-[CM] mRNA accumulation at 1, 2, 3, and 4 wpi in infected N. benthamiana and 2, 3, 4, and 5 wpi in infected cassava (B) and of EACMCV viral mRNA accumulation at 2, 3, 4, and 5 wpi in infected N. benthamiana and cassava (E). Total RNA (2 μg) isolated from virus-infected plants was loaded into each lane. Blots were hybridized with [∝-32P]dCTP-labeled ACMV-[CM]-specific (A and B) and EACMCV-specific (D and E) probes. Ethidium bromide-stained rRNA was shown as the loading control. (C and F) Correlation of viral DNA and mRNA accumulation in ACMV-[CM]-infected (C) and EACMCV-infected (F) N. benthamiana and cassava plants.