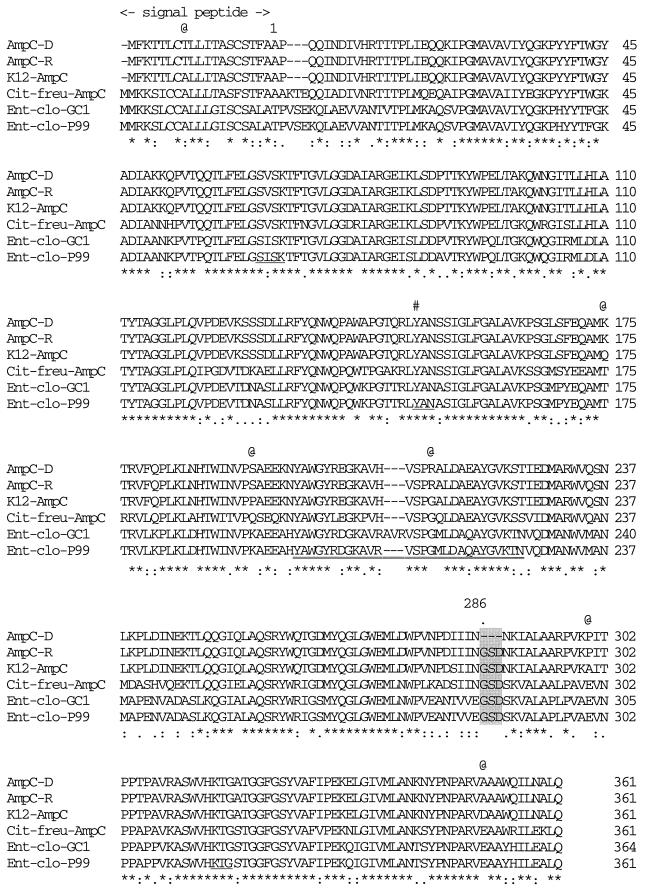
FIG. 1.
Predicted amino acid sequence of the AmpC β-lactamase of E. coli HKY28 aligned with that of E. coli K-12 (10). The 3-amino-acid deletion in the HKY28 AmpC is shaded. Underlines, the β-lactamase active site SVSK, the conserved tripeptide KTG, and the class C motif YXN; #, position of Tyr150; @, positions of the amino acid substitutions observed between the AmpCD of strain HKY28 and the AmpC of strain K-12; numbers on the right, numbers of amino acid residues from the N terminus of each mature protein; ✻, amino acid residues conserved among the six AmpC-type enzymes; colons and dots, amino acid substitutions that result in homologous amino acid residues; Cit-freu, Citrobacter freundii; Ent-clo, E. cloacae; double underline, AmpC Ω-loop domain.