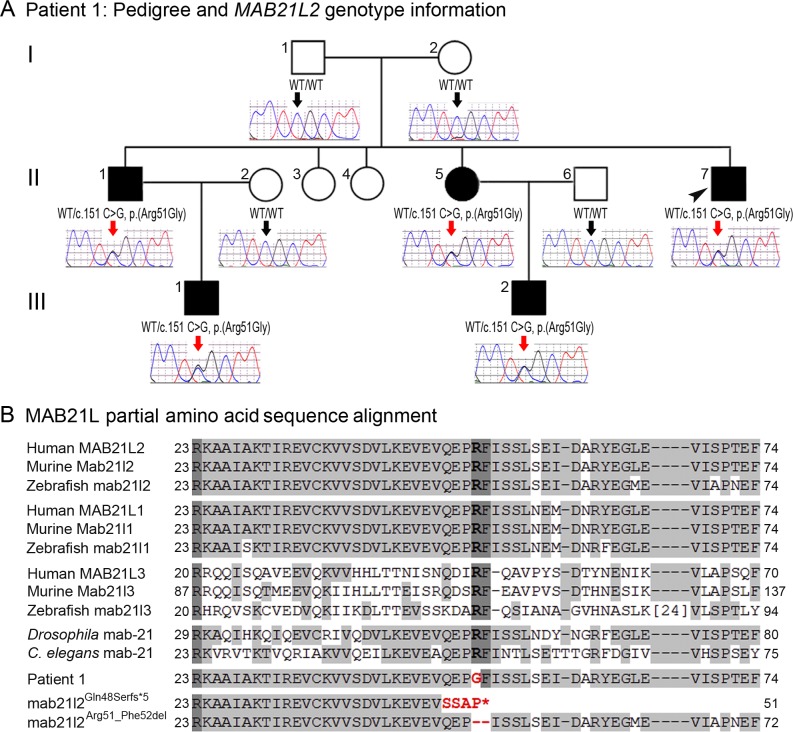
Fig 2. MAB21L2 mutations and protein sequence conservation.
A. Three-generation pedigree of Patient 1 with MAB21L2 genotype information. DNA chromatograms for all tested family members are shown with c.151C position indicated with black (WT allele) or red (heterozygous mutant allele) arrows. The proband (Patient 1) is indicated with a black arrowhead. Please note the presence of the mutant allele in all affected individuals, its absence in unaffected family members, and the presence of a low ‘G’ peak in addition to the normal ‘C’ nucleotide at the mutant position in the proband’s unaffected mother. B. Amino acid alignment of the MAB21L1-3 and mab-21 regions surrounding the arginine at position 51; amino acids identical between different homologs are highlighted with a light grey color, three invariant residues are shown in dark grey; the glycine (G) predicted to replace arginine 51 in Patient 1 is shown in red font; the positions and predicted effects of the zebrafish mab21l2 mutations involving the same region are also shown in red font. Accession numbers for sequences utilized in the alignment are provided in Methods.