Abstract
Background
Glucosamine and chondroitin are popular non-vitamin dietary supplements used for osteoarthritis. Long-term use is associated with lower incidence of colorectal and lung cancers and with lower mortality; however, the mechanism underlying these observations is unknown. In vitro and animal studies show that glucosamine and chondroitin inhibit NF-kB, a central mediator of inflammation, but no definitive trials have been done in healthy humans.
Methods
We conducted a randomized, double-blind, placebo-controlled, cross-over study to assess the effects of glucosamine hydrochloride (1500 mg/d) plus chondroitin sulfate (1200 mg/d) for 28 days compared to placebo in 18 (9 men, 9 women) healthy, overweight (body mass index 25.0–32.5 kg/m2) adults, aged 20–55 y. We examined 4 serum inflammatory biomarkers: C-reactive protein (CRP), interleukin 6, and soluble tumor necrosis factor receptors I and II; a urinary inflammation biomarker: prostaglandin E2-metabolite; and a urinary oxidative stress biomarker: F2-isoprostane. Plasma proteomics on an antibody array was performed to explore other pathways modulated by glucosamine and chondroitin.
Results
Serum CRP concentrations were 23% lower after glucosamine and chondroitin compared to placebo (P = 0.048). There were no significant differences in other biomarkers. In the proteomics analyses, several pathways were significantly different between the interventions after Bonferroni correction, the most significant being a reduction in the “cytokine activity” pathway (P = 2.6 x 10-16), after glucosamine and chondroitin compared to placebo.
Conclusion
Glucosamine and chondroitin supplementation may lower systemic inflammation and alter other pathways in healthy, overweight individuals. This study adds evidence for potential mechanisms supporting epidemiologic findings that glucosamine and chondroitin are associated with reduced risk of lung and colorectal cancer.
Trial Registration
ClinicalTrials.gov NCT01682694
Introduction
Glucosamine and chondroitin (G&C), often taken together as a single pill and used for osteoarthritis, are among the most popular dietary supplements in the US [1]. G&C are considered safe, with no known major adverse side effects [2]. While the efficacy of these supplements for treatment of osteoarthritis remains debated [2,3], recent studies suggest that they may have potential for reducing risk of other chronic diseases, such as cancer. In the VITamins And Lifestyle (VITAL) study, a large (n = 77,738) prospective cohort, we reported that use of G&C supplements is associated with a 27–35% lower incidence of colorectal cancer [4], a 26–28% lower incidence of lung cancer [4], 17% lower overall mortality [5], and a 13% lower cancer mortality [6]. These findings on G&C were the most consistent results across endpoints among the 30 supplements that were the focus of the VITAL study.
It is generally well-accepted that chronic inflammation contributes to carcinogenesis [7]. The majority of cellular events involved in the inflammation process require nuclear factor kappa B (NK-κB), a transcription factor that plays a central role in the generation of cytokines, chemokines and other soluble factors involved in the immune response. Similarly, excess reactive species characteristic of oxidative stress can damage cell membranes and DNA [8], and are thought to contribute to genomic instability and development of cancer [9]. Several lines of compelling evidence from in vitro and preclinical studies support a possible role for G&C in reducing inflammation [10–13]. A limited number of human observational and randomized studies also suggest that G&C lower systemic biomarkers of inflammation and oxidative stress [14,15]; however, no human intervention trials have evaluated the potential for G&C to reduce inflammation or alter other pathways relevant to carcinogenesis in healthy individuals free of chronic inflammatory conditions.
Our objectives in the present study were two-fold. Our first aim was to determine the effect of a common dose of G&C on a panel of serum inflammation biomarkers [high sensitivity C-reactive protein (CRP), interleukin-6 (IL-6), and soluble tumor necrosis factor receptors I & II (sTNFRI and II); a urinary biomarker of inflammation, prostaglandin E2-metabolite (PGE-M); and a urinary biomarker of oxidative stress, F2-isoprostane (F2-IsoP)]. Secondly, we sought to characterize intervention-induced changes in plasma proteomic patterns via pathway analyses in response to G&C compared to placebo. Results from this study may help determine the anti-inflammatory potential of G&C in humans, and may identify other possible pathways that may be modulated with use of G&C.
Methods
Research Design
The study was a randomized, double-blind, placebo-controlled, crossover trial comparing supplemental glucosamine and chondroitin to placebo. The protocol for this trial and supporting CONSORT checklist are available as supporting information; see S1 Protocol and S1 CONSORT Checklist. As this was a small pilot trial with the aim of enrolling a sample of 20 individuals, participants were randomly assigned by the project manager to begin with either the active or placebo intervention period in blocks of two using a pre-determined randomization template alternating placebo and G&C interventions. Each new recruit was assigned to the next available assignment. Each intervention period lasted 28 days with a 28 day washout period between the two interventions. All study activities were carried out at the Fred Hutchinson Cancer Research Center (FHCRC), Seattle, Washington. Recruitment, enrollment, trial, and sample collection took place from October 2012 to July 2013. The study protocol was approved by the FHCRC Institutional Review Board on July 11, 2012, with continuation approval obtained through May 18, 2015. All participants gave informed written consent. The authors confirm that all ongoing and related trials for this intervention are registered. This trial was registered at http://www.clinicaltrials.gov as NCT01682694.
Participants
Participants were healthy, overweight [BMI (body mass index) >25 and ≤32.5 kg/m2], non-smoking, aged 20–55 years, and recruited from the greater Seattle area. We recruited men and women who were overweight because inflammation biomarkers are unlikely to decrease measurably in response to an intervention among individuals who have very low systemic inflammation at baseline, and BMI is a strong predictor of both CRP and IL-6 [16,17]. Methods for recruitment included flyers posted on university campuses in Seattle and at FHCRC, an informational website, and advertisements in campus and local neighborhood newspapers.
Screening occurred in two phases. In the first phase, individuals were screened for eligibility using a self-administered questionnaire and were excluded for any of the following: chronic medical illness; history of gastrointestinal, hepatic, or renal disorders; inflammatory conditions (including autoimmune and inflammatory diseases); pregnancy or lactation; currently on a weight loss diet; BMI <25 or ≥32.5 kg/m2; alcohol intake >2 drinks/day (2 drinks defined as 720 ml beer, 240 ml wine or 90 ml spirits); current use of prescription or over-the-counter medications other than oral contraceptives and hormone-secreting IUDs, multivitamins or use of aspirin or NSAIDs more than 2 days per week; inability to swallow pills; known allergy to shellfish; not willing to take pills made from shellfish or animal sources; or intention to relocate out of the study area within the next 4 months. Eligible individuals were invited to a study information session and those interested in participating provided informed written consent.
In the second phase of screening, prospective participants attended a screening clinic visit at the FHCRC Prevention Center. Height and weight were measured to ensure BMI requirements were met. Because body fat can vary among similar BMIs, body fat percentage was assessed by whole-body dual-energy X-ray absorptiometry (DEXA) scanning using a GE Lunar DPX-Pro densitometer (GE Healthcare, Milwaukee, WI). Blood was drawn in the morning after an overnight fast and was used for analysis of renal (blood urea nitrogen: 7–25 mg/dl; creatinine: 0.50–1.10 mg/dl for females; 0.60–1.35 mg/dl for males; eGFR ≥ 60 ml/min/1.73m2; sodium: 135–146 μmol/l; potassium: 3.5–5.3 μmol/l; chloride: 98–110 μmol/l), liver (albumin: 3.6–5.1 g/dl; globulin: 2.1–3.7; total bilirubin: 0.2–1.2 mg/dl; alkaline phosphatase: 40–115 U/L; AST: 10–30 U/L for females; 10–40 U/L for males; ALT: 6–40 U/L for females; 9–60 U/L for males), and metabolic function (glucose: 65–99 mg/dl). Individuals with normal laboratory values were invited to participate in the study.
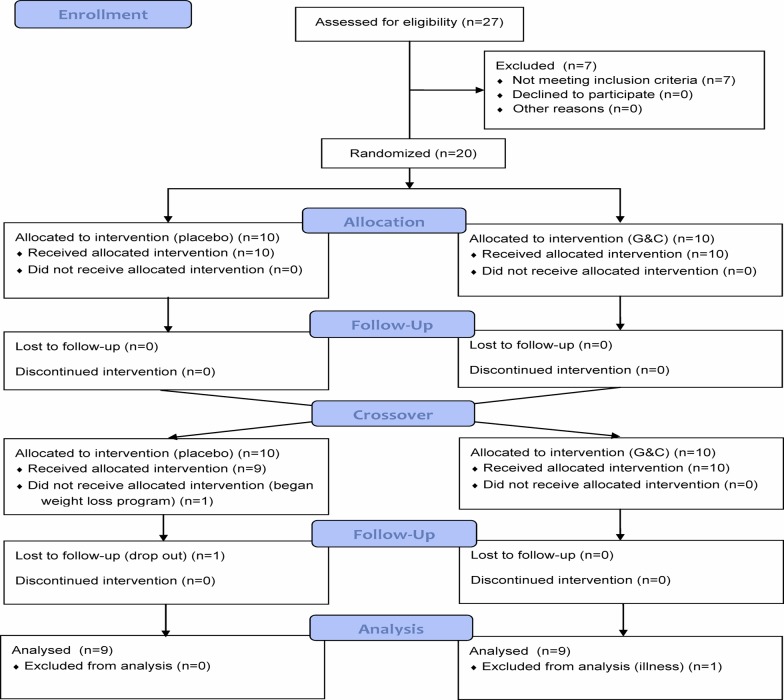
A total of 27 individuals underwent screening activities. Seven were ineligible prior to randomization, six due to high fasting glucose concentrations and one due to abnormal liver enzyme concentrations. Twenty individuals (n = 10 men; n = 10 women) were randomized. One female was asked to drop out after completing only one intervention period because she started a new diet and exercise program that might have had effects on biomarker measures. Because inflammation biomarkers are influenced by minor infections (e.g., colds) and other illnesses [18], participants were instructed to delay clinic visits until they were no longer ill; however, one male participant attended a clinic visit at the onset of a cold and was also excluded, leaving a sample size of 18 (9 men and 9 women; Fig. 1).
Fig 1. CONSORT Flow diagram of participants in the glucosamine and chondroitin (G&C) randomized, placebo-controlled trial.
Glucosamine and Chondroitin Supplements
Our aim was to evaluate G&C as used in population settings. Thus, dosing was based on the common starting dose recommended by the product manufacturer, which is similar across G&C products. The active treatment was 1500 mg/d FCHG49 glucosamine hydrochloride (GHCl) + 1200 mg/d TRH122 sodium chondroitin sulfate (CS) taken as 3 capsules daily with each capsule containing 500 mg GHCl and 400 CS. Crystalline cellulose was used as an inactive filler, and both active treatment and placebo were encapsulated in clear gelatin capsules. The placebo was similar in appearance and contained only the inactive filler. Both G&C and the placebo were generated from a single lot of source materials and donated by Nutramax Laboratories Consumer Care, Inc. (Edgewood, MD). Supplement bottles were provided by the manufacturer with a single printed label containing the letters “A” or “B”. To maintain double-blinding for both participants and investigators, the randomization algorithm was sent in a sealed envelope to an investigator uninvolved in the day to day study activities; unblinding of the interventions was done at the time of data analysis. Participants were contacted by study staff at the mid-point and end of each intervention period to assess any possible adverse experiences and compliance. No adverse events were reported. Extra pills were included in each supplement bottle in the event that a Day 28 study visit was delayed, and so that adherence percentages (pills supplied—pills returned/days elapsed) could be calculated as an additional compliance measure. Composition of analysis testing was performed by an independent laboratory (Tampa Bay Analytical Research, Inc., Clearwater, FL) in collaboration with the NIH Office of Dietary Supplements. HPLC-UV analysis of a subset of capsules confirmed that mean G&C content was within specifications. No G&C was detected in the placebo capsules and no microbial products were detected in either capsule type.
Specimen Collection
Biological samples were collected at baseline (screening visit) and after each 28-day intervention period in the morning after a minimum of a 12-hour overnight fast. For blood samples, tubes without additive were allowed to clot at room temperature for 30 minutes before they were centrifuged to separate the serum. Blood was also drawn into a tube containing EDTA for plasma. Both serum and plasma were aliquoted and stored at -80°C.
Participants were given a urine collection kit and instructions before each visit, and brought the specimen with them to the clinic visit. Participants were instructed to collect all of their urine for the 12 hours prior to their morning clinic visit at baseline and on Day 28 of each intervention period. Urine-void times were recorded and any uncollected voids noted. Urine specimens were stored at 4°C until delivery to the FHCRC the morning following collection. Urine volume was measured and aliquots stored at 80°C.
Laboratory Analyses
Serum Inflammation Biomarkers
All assays were performed on never-thawed samples with the exception of CRP which is stable through freeze-thaw cycles, and was measured in samples that had been thawed once [19]. No observations were below the limit of detection for any markers. Assays for all inflammation markers have been described [20]. Intra and inter-assay CVs for CRP, IL-6, and sTNFRI and II were 1.7% and 5.5%; 3.7% and 5.8%; 2.1% and 1.7%; and 2.3% and 5.6%, respectively.
Urinary Inflammation and Oxidative Stress Biomarkers
Urinary PGE-M (11-alpha-hydroxy-9,15-dioxo-2,3,4,5-tetranor-prostane-1,20-dioic acid), the stable metabolite of PGE2, was assayed at the Vanderbilt Eicosanoid Core Laboratory (VECL, Nashville, TN) by a liquid chromatography/mass spectrometry (LC/MS) method, as previously described [21]. Urinary F2-IsoP was measured by gas-chromatography (GC)-negative-ion chemical ionization MS by the VECL using an Agilent 5973 GC/MS system (Santa Clara, CA) [22].
Proteomics Analysis of Plasma
Plasma samples were evaluated on a customized antibody array populated with ~3,000 full-length antibodies, printed in triplicate on a single microarray [23–25]. Briefly, to analyze proteins in the plasma samples, we depleted albumin and IgG. Protein (200 μg) from the two participant blood draws were each labeled with Cy5. Each sample was combined with reference sample and pipetted at the microarray/coverslip junction followed by incubation for 90 min at room temperature in the dark. Unbound proteins were removed by washing and the slides scanned for Cy3 and Cy5 fluorescence in an Axon Genepix 4000B scanner. The Cy5/Cy3 ratio determined the relative concentration of protein compared to reference. The array contains antibodies to cytokines (e.g., interleukins 1A, 1B, 2, 3, 4, 4R, 5, 6, 7, 8, 10, 12A, 13, 15, 16, 17D, 18, 19, 20, 24, 28A, and 34 and interleukin receptors 2RA, 2RA, 3RA, 4R, 5RA, 6ST, 17RA and 17RB), adipokines, proteins involved in apoptosis, angiogenesis, T-cell activation/infiltration, inflammation/prostaglandins, insulin, insulin resistance toll-like receptor (TLR), transforming growth factor (TGF)-β, and STAT signaling pathways that have been reported to be deregulated in a variety of diseases. Most (>85%) antibodies on the array had coefficients of variation, for triplicates, of less than 10% [23,24,26,27]. Detailed protocol descriptions of array fabrication, plasma treatment, plasma labeling, incubation, and scanning have been published elsewhere [23,27].
Array Analysis and Normalization
Array data contain a format identical to two-channel gene expression arrays and analysis proceeds analogously. The array image was scanned using a GenePix 4000B (Axon Instruments) scanner. The numerical data processed by Genepix Pro 6.0 were imported to Limma 2.4.11 (Linear Models for Micro Array, a Bioconductor R package [28]). For each antibody, fold-change of the signal (red channel) was compared to the reference (green channel). The M value was calculated as Red/Green, where Red and Green were computed after background correction using the “normexp” method [29]. Saturated array spots were flagged and triplicate antibodies with coefficients of variation >10% were removed. Experimental variation was normalized using within-array print-tip loess and between-array quartile normalization. Triplicate features were summarized using their median. M values were standardized such that the mean value and standard deviation of the placebo group was set to zero and one, respectively. After all processing, data were available for analysis on a total of 2938 antibodies.
Statistical Analysis
All inflammatory and oxidative stress biomarkers were log-transformed to improve the normality of the distributions prior to analysis. Results are presented as geometric means with 95% confidence intervals (CI). Generalized estimating equations (GEE), which account for correlation of repeated measures, were used to assess the effects of G&C versus placebo on inflammation and oxidative stress. Analysis of each Day 28 inflammatory or oxidative stress biomarker was adjusted for the same biomarker concentration at Day 0 for each treatment period. Sex, age, and body-fat percent were selected a priori as additional covariates for adjustment in the model. We also evaluated the effect of treatment period but because it did not affect the point estimates, (i.e., carryover effects were not contributing to outcome measures), we did not include treatment period in the models. The two-sided P value for statistical significance for biomarkers of inflammation and oxidative stress was set at <0.05. Analyses were performed using Stata statistical software (v13.1, StataCorp, College Station, TX).
Proteomics data were analyzed by performing paired t-tests to determine statistically significant differences between the two interventions, after adjusting for covariate effects, including hybridization day, and plate and box position by linear regression. Antibody markers were ranked on the basis of p value and effect sizes, so that a positive value indicates that the antibody expression was greater after supplementation with G&C compared to placebo, and a negative value indicates lower expression. To further integrate the antibody at the protein level, multiple duplicated antibodies for the same protein that did not have at least two significant results with the effect size going in the same direction were excluded. Among those proteins with at least two concordant significant results, the most significant antibody was reported for the top 100 proteins. Gene Set Enrichment Analyses, the evaluation of microarray data at the level of “gene groups” that share a common biologic theme or cellular location, were carried out based on the t-test results using Kyoto Encyclopedia of Genes and Genomes (KEGG) [30] and the gene set database Gene Ontology (GO) [31] downloaded from the Molecular Signatures Database (MSigDB, http://www.broadinstitute.org/gsea/msigdb/index.jsp). We used a Bonferroni correction to determine significant differences within the KEGG pathway [P <3.85x10-4; (0.05/130, the total number of pathways evaluated in KEGG)], the GO pathway [P<5.18x10-5; (e.g., 0.05/966, the total number of pathways evaluated in GO)], and for the individual proteins [P <1.70x10-5; (0.05/2,938, the total number of antibodies on the array)].
Results
Table 1 summarizes the demographic and baseline characteristics of the 18 study participants stratified by sex. Baseline measures of all inflammatory biomarkers were higher among men, while F2-isoP was lower.
Table 1. Characteristics of participants randomized in the crossover trial of glucosamine and chondroitin at baseline (n = 18).
| Men (n = 9) | Women (n = 9) | |
|---|---|---|
| Personal characteristics 1 | ||
| Age (y) | 40 (8.0) | 38 (8.8) |
| BMI (kg/m 2 ) | 27 (2.8) | 29 (2.4) |
| Body fat (%) 2 | 32 (4.6) | 39 (4.1) |
| Inflammation biomarkers 3 | ||
| CRP mg/l | 1.18 (1.3) | 0.77 (1.1) |
| IL-6 pg/ml | 1.19 (0.9) | 0.52 (0.2) |
| sTNFRI pg/ml | 1119 (287) | 654 (398) |
| sTNFRII pg/ml | 5832 (1280) | 5398 (620) |
| PGE-M (ng/mg creatinine) | 8.32 (4.7) | 3.83 (1.3) |
| F2-isoprostane (ng/mg creatinine) | 1.00 (0.4) | 1.13 (0.4) |
1Mean (SD)
2Measured by Dual X-ray Absorptiometry (DEXA)
3Geometric mean (geometric SD)
Statistically significantly lower geometric mean CRP concentrations were observed after the G&C intervention compared to placebo (-23%, P = 0.048; Table 2). No significant differences were observed in the other inflammation or oxidative stress biomarkers assessed, although they were slightly lower after G&C supplementation with the exception of the soluble TNF receptors, which increased by 1–3%.
Table 2. Inflammatory and oxidative stress biomarker concentrations after placebo and glucosamine and chondroitin intervention.
| Biomarker | Placebo (N = 18) | Glucosamine & Chondroitin (N = 18) | P |
|---|---|---|---|
| Mean (SD) 1 | Mean (SD) 1 | ||
| CRP (mg/l) | 1.17 (0.17) | 0.90 (0.13) | 0.048 |
| IL-6 (pg/ml) | 0.89 (0.10) | 0.81 (0.09) | 0.27 |
| sTNFRI (pg/ml) | 871.3 (15.6) | 901.6 (15.7) | 0.17 |
| sTNFRII (pg/ml) | 5558 (103) | 5633 (104) | 0.34 |
| PGE-M (ng/mg creatinine) | 6.15 (0.41) | 5.89 (0.39) | 0.60 |
| F2-isoprostane (ng/mg creatinine) | 1.20 (0.08) | 1.10 (0.08) | 0.38 |
1Least squares geometric means (geometric SD) from GEE model adjusted for baseline values, age, sex, and body fat percent
Of the ~3,000 antibodies on the proteomics array, 2,938 were detected. We conducted gene-set enrichment pathway analyses using both KEGG and GO database tools (Table 3). Five pathways in the KEGG pathway analysis remained statistically significant after a Bonferroni correction (nominal p-values 7 x 10-10 to 4 x 10-5)—cytokine-cytokine receptor interaction, JAK/STAT signaling pathway, intestinal immune network for IGA production, leukocyte transendothelial migration, and ubiquitin-mediated proteolysis. Twenty-five pathways in the GO pathway analyses remained statistically significant after Bonferroni correction (p values 3 x 10-16 to 1 x 10-5), including cytokine activity, receptor binding, hematopoietin/interferon (classD 200 domain) cytokine receptor binding, growth factor activity, extracellular region, chemokine activity, extracellular space, G-protein coupled receptor binding, chemokine receptor binding, extracellular region part, locomotory behavior, membrane fraction, response to external stimulus, regulation of actin filament length, regulation of actin polymerization and/or depolymerization, regulation of cellular component size, behavior, actin polymerization and/or depolymerization, regulation of organelle organization and biogenesis, regulation of cytoskeleton organization and biogenesis, response to biotic stimulus, nucleus, microtubule-based process, response to other organism and cell fraction. In each analysis, cytokine activity/cytokine receptor binding was the most significantly affected pathway. With the exception of ubiquitin-mediated proteolysis, and proteins located in the nucleus, all pathways were down-regulated after G&C supplementation.
Table 3. Significantly different pathways comparing glucosamine and chondroitin supplementation to placebo via gene-set enrichment analysis in Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases (n = 18).
| Pathway/ Genes (count) 1 | Probes in Array 2 | Positive Probes 3 | Negative Probes 4 | P value* | Genes in the Pathway |
|---|---|---|---|---|---|
| KEGG 5 | |||||
| Cytokine-Cytokine Receptor Interaction | |||||
| 101 | 368 | 70 | 165 | 7.2x10-12 | CSF3;IL13;IL8;EGFR;TNF;LIFR;CXCL12;PRLR;CCL20;IL4;IL6;IL10;CCL5;TNFRSF9;PRL;FLT4;IL5RA; CD40LG;IL28A;KIT;VEGFA;IL24;TNFRSF17;LEP;CD27;IFNGR1;IL15;IFNB1;CCL14;TNFRSF1B;IL20; IL15RA;FLT3;FASLG;IL18;CSF2;MPL;IL1A;IL1RAP;IL18RAP;PDGFRA;CSF1;LEPR;TNFRSF11B; TNFRSF10B;IL2RA;NGFR;IL10RB;FIGF;CXCL11;CSF3R;IFNG;TGFB1;CXCR4;PF4;CCR5;IL6ST; TGFBR2;IL3RA;CCL4;TGFB3;IFNAR1;IL12A;TNFSF8;PDGFRB;IL4R;MET;EGF;IL7;TGFB2;KDR;CXCL5;TNFSF13B;CCL27;IL5;FLT1;IL13RA1;CCR7;TNFSF9;CXCR3;BMPR1A;IL19;TNFRSF10A;TNFRSF8; FAS;INHBE;PDGFA;BMP7;INHBC;CCL21;CCR2;GHR;CCR6;PDGFB;VEGFB;OSM;CD70;CXCL10; IFNA1;IL12RB2;INHBA |
| JAK/STAT Signaling | |||||
| 71 | 211 | 39 | 104 | 4.4x10-8 | CSF3;IL13;LIFR;PRLR;IL4;IL6;IL10;CCND3;PRL;IL5RA;IL28A;IL24;LEP;IFNGR1;IL15;IFNB1;PIK3CB; STAT5A;CCND1;SOCS7;MYC;IL20;IL15RA;CSF2;MPL;PIM1;CBLB;LEPR;STAT1;IL2RA;IL10RB;CSF3R;IFNG;CSH1;GRB2;PIK3CA;IL6ST;IL3RA;IFNAR1;IL12A;AKT1;IL4R;IL7;PTPN6;STAT5B;PTPN11;IL5; STAT4;IL13RA1;BCL2L1;SOCS2;SOS1;STAT3;IL19;CCND2;SPRED1;AKT2;CBL;JAK3;TYK2;GHR; SOCS1;SPRY1;PIK3R1;CREBBP;OSM;PIK3CD;IRF9;CBLC;IFNA1;IL12RB2 |
| Intestinal Immune Network for IGA Production | |||||
| 15 | 88 | 17 | 51 | 8.7x10-6 | CXCL12;IL4;IL6;IL10;CD40LG;CD86;TNFRSF17;IL15;IL15RA;TGFB1;CXCR4;CD28;TNFSF13B;CCL27; IL5 |
| Leukocyte Transendothelial Migration | |||||
| 38 | 109 | 19 | 53 | 3.5x10-5 | CXCL12;CLDN18;ITGB1;PTK2B;RAC1;MMP9;PLCG1;PIK3CB;THY1;VCL;BCAR1;VASP;MMP2; VCAM1;ACTB;PECAM1;RAP1A;RHOA;PTK2;CXCR4;PIK3CA;ITGAL;ITK;GNAI3;PTPN11;VAV3;PXN; CDH5;CLDN6;RASSF5;PRKCA;PIK3R1;MAPK11;PIK3CD;CLDN1;ITGAM;SIPA1;MAPK14 |
| Ubiquitin-Mediated Proteolysis | |||||
| 23 | 37 | 19 | 3 | 4.1x10-5 | BRCA1;MDM2;BIRC2;BIRC3;MAP3K1;CBLB;CUL4A;CUL2;CUL5;XIAP;CUL4B;CUL7;UBE2S;FANCL; CUL1;CBL;UBR5;SOCS1;ANAPC2;SKP2;VHL;FBXW7;CBLC |
| GO 5 | |||||
| Cytokine Activity | |||||
| 41 | 177 | 23 | 109 | 2.6x10-16 | CSF3;IL8;TNF;CXCL12;CCL20;IL4;CCL5;PRL;CD40LG;VEGFA;BMP4;ERBB2;IL20;CSF2;CSF1; TNFRSF11B;FIGF;CXCL11;IFNG;C5;IL1RN;PF4;CCL4;IL12A;IL7;TGFB2;TRIP6;CXCL5;CCL27;IL5;IL19; INHA;SPRED1;GDF15;CCL21;PIK3R1;OSM;CXCL10;MUC4;MIF;INHBA |
| Receptor Binding | |||||
| 109 | 280 | 48 | 151 | 2.5x10-14 | CSF3;HBEGF;IL8;EFNB3;BID;IRS1;TNF;CXCL12;CCL20;EREG;IL4;CCL5;LCK;NCK1;DLL4;ANXA1; NCOR2;PRL;NMB;CD40LG;TGFA;FBLN5;SLC9A3R1;VEGFA;ICAM2;GRN;NCOA6;EFNB1;THY1; BMP4;STOML2;ERBB2;IL20;CSF2;ANG;CSF1;PCSK9;ALCAM;FGF1;TNFRSF11B;CYTL1;VWF;GAST; FIGF;F2R;ADAM23;ADAM9;CXCL11;IFNG;COL4A3;TGFB1;GFRA1;IGF1;C5;GRB2;NPY;MED12; PTHLH;MBL2;EDN1;IL1RN;GABARAP;AREG;ADAMTS13;STC2;DLL1;PF4;CCL4;EFNA5;TGFB3; IL12A;EGF;IL7;TGFB2;TRIP6;CXCL5;CCL27;FGF3;IL5;WNT5A;SOCS2;FADD;TGFB1I1;F2;IL19; NUP62;INHA;TNXB;JMJD1C;GABARAPL2;SPRED1;GDF15;INHBC;RLN1;CCL21;CCR2;CHGB;SOCS1;DST;PIK3R1;OSM;CXCL10;MUC4;TDGF1;CASP8AP2;MIF;JAG2;SHC1;INHBA |
| Hematopoietin/Interferon Cytokine Receptor Activity | |||||
| 8 | 68 | 5 | 48 | 1.7x10-12 | CSF3;IL4;PRL;CSF2;IFNG;IL7;IL5;OSM |
| Growth Factor Activity | |||||
| 18 | 79 | 8 | 51 | 1.0x10-10 | CSF3;IL4;TGFA;GRN;CSF2;CSF1;FGF1;IL1RN;AREG;EGF;IL7;TRIP6;FGF3;IL5;INHA;TDGF1;JAG2; INHBA |
| Extracellular Region | |||||
| 140 | 293 | 58 | 145 | 1.3x10-8 | THBS4;CSF3;HBEGF;COMP;IL8;DSPP;CCL20;MMP3;EREG;IL4;IL6;TFF3;PLA2G5;CTGF;IL5RA;CPN1;CD5L;DGCR6;ORM1;FBLN5;PNLIPRP2;MMP9;VEGFA;DCD;S100A7;COL18A1;LEP;TFRC;IL15; CCL14;MMP2;PSAP;ERBB2;IL20;SERPINA5;IL18;LGALS3BP;ANG;IL1A;GSN;MMP7;PCSK9;SOD1; CDA;PRG2;CDH13;TNFRSF11B;CYTL1;CP;SFRP4;VWF;COL8A1;LGALS7;TPT1;SERPINA1;FIGF; CFHR1;SPINT2;FBN1;COL4A3;TGFB1;PI3;IL16;LOXL1;KLK5;C5;FGG;WNT2B;NPY;PTHLH;MBL2; EDN1;IL1RN;AREG;ADAMTS13;SFRP1;SFN;PRSS8;MMP13;CCL4;EFNA5;IL12A;TINAG;IL7;TGFB2; SHH;LAMC1;IGFBP1;FGF3;FRZB;MMP10;IL5;WNT5A;DKK3;KLK10;APCS;F7;MMP11;FLT1;APOD; LAMB1;ANGPTL4;CRLF1;CPB2;FBLN2;POSTN;PDZD2;LYZ;KLK6;MUC5AC;NUCB1;F2;C2;MSMB; INHA;FSTL1;TNXB;F13A1;PTX3;GDF15;KLK8;VTN;INHBC;KLK13;COL5A1;COL5A3;PLG;DST;OSM; FGF2;RBP4;ADM;A1BG;FGFBP1;LALBA;REG3A;FBN2;MIF;DMBT1;INHBA |
| Chemokine Activity | |||||
| 12 | 70 | 9 | 49 | 1.7x10-8 | IL8;CXCL12;CCL20;CCL5;CXCL11;C5;PF4;CCL4;CXCL5;CCL27;CCL21;CXCL10 |
| Extracellular Space | |||||
| 83 | 196 | 37 | 105 | 2.2x10-8 | CSF3;HBEGF;IL8;CCL20;MMP3;EREG;IL4;IL6;IL5RA;CPN1;CD5L;ORM1;MMP9;VEGFA;LEP;IL15; CCL14;MMP2;PSAP;LGALS3BP;ANG;IL1A;MMP7;PCSK9;SOD1;CDH13;CYTL1;CP;SFRP4;LGALS7; TPT1;SERPINA1;FIGF;CFHR1;FBN1;TGFB1;IL16;KLK5;C5;FGG;WNT2B;NPY;PTHLH;MBL2;EDN1; IL1RN;AREG;SFRP1;SFN;PRSS8;MMP13;CCL4;EFNA5;IL12A;SHH;IGFBP1;MMP10;IL5;WNT5A; DKK3;APCS;FLT1;APOD;CRLF1;CPB2;LYZ;NUCB1;F2;C2;MSMB;INHA;FSTL1;KLK8;VTN;PLG;OSM; FGF2;RBP4;ADM;FGFBP1;LALBA;REG3A;INHBA |
| G-Protein-Coupled Receptor Binding | |||||
| 14 | 75 | 10 | 51 | 2.3x10-8 | IL8;CXCL12;CCL20;CCL5;SLC9A3R1;CXCL11;C5;PF4;CCL4;CXCL5;CCL27;CCL21;CCR2;CXCL10 |
| Chemokine Receptor Binding | |||||
| 13 | 72 | 9 | 49 | 2.4x10-7 | IL8;CXCL12;CCL20;CCL5;CXCL11;C5;PF4;CCL4;CXCL5;CCL27;CCL21;CCR2;CXCL10 |
| Extracellular Region | |||||
| 106 | 227 | 46 | 118 | 7.3x10-8 | THBS4;CSF3;HBEGF;COMP;IL8;DSPP;CCL20;MMP3;EREG;IL4;IL6;CTGF;IL5RA;CPN1;CD5L;DGCR6;ORM1;FBLN5;MMP9;VEGFA;COL18A1;LEP;IL15;CCL14;MMP2;PSAP;LGALS3BP;ANG;IL1A;MMP7;PCSK9;SOD1;CDH13;CYTL1;CP;SFRP4;COL8A1;LGALS7;TPT1;SERPINA1;FIGF;CFHR1;FBN1; COL4A3;TGFB1;PI3;IL16;KLK5;C5;FGG;WNT2B;NPY;PTHLH;MBL2;EDN1;IL1RN;AREG;ADAMTS13;SFRP1;SFN;PRSS8;MMP13;CCL4;EFNA5;IL12A;TINAG;SHH;LAMC1;IGFBP1;MMP10;IL5;WNT5A; DKK3;APCS;MMP11;FLT1;APOD;LAMB1;CRLF1;CPB2;FBLN2;POSTN;LYZ;MUC5AC;NUCB1;F2;C2;MSMB;INHA;FSTL1;TNXB;KLK8;VTN;COL5A1;COL5A3;PLG;DST;OSM;FGF2;RBP4;ADM;FGFBP1; LALBA;REG3A;FBN2;INHBA |
| Locomotory Behavior | |||||
| 33 | 112 | 17 | 64 | 2.4x10-7 | IL8;CXCL12;CCL20;IL4;IL10;CCL5;PIK3CB;SOD1;MAPK1;FOSL1;CDH13;PLD1;CXCL11;C5;CXCR4; DEFA1;PF4;CCR5;TGFB2;CXCL5;CCL27;MAP2K1;CCR7;CXCR3;PLAUR;RALBP1;CCL21;CCR2;CCR6; FGF2;HRAS;CXCL10;MAPK14 |
| Membrane Fraction | |||||
| 42 | 113 | 17 | 59 | 2.5x10-7 | CEACAM1;VCP;BID;IRS1;EEA1;GALK1;LYVE1;IL15;BCAR1;FOLR3;FUT3;LY6D;ATP8B1;STX16;JUP; FUT4;STEAP2;CD59;HLAB;GP1BA;PRKCE;DPP10;APOB;XRCC6;SPTAN1;ABCB1;DAG1;CLIC1;CDH5;PTGS1;ABCA3;SLC1A1;LRP12;AMFR;PRKCA;SELP;PRKCZ;CD70;LRP1;PDLIM5;FOLH1;SLC2A1 |
| Response to External Stimulus | |||||
| 108 | 249 | 47 | 122 | 2.9x10-7 | IL13;IL8;ITGB3;CXCL12;WAS;CCL20;EREG;IL4;IL10;CCL5;FOS;CD97;F13B;MMRN1;PLAT;ANXA1; CTGF;CD40LG;ORM1;RPS6KA5;RAC1;S100A8;LEP;LYVE1;SSTR2;F5;PIK3CB;MDK;SELE;IL20;TFPI; IL1A;IL1RAP;IL18RAP;PCSK9;SOD1;MAPK1;FOSL1;CDH13;S100A9;CD59;VWF;IL10RB;PLD1; CFHR1;F2R;GP1BA;GCGR;CXCL11;TGFB1;TP53;C5;CHST2;NMI;CXCR4;NPY;MBL2;DEFA1;STC2; PF4;CCR5;MGLL;CCL4;CRP;CDKN1A;CHMP1A;TNFAIP6;TGFB2;CXCL5;CCL27;NF1;IL5;APCS;F7; MAP2K1;CCR7;RTN4RL2;LYZ;CXCR3;PPARG;CDKN2B;F2;S100A12;AOAH;C2;SERPINE1;INHA; F13A1;PLAUR;PTX3;KLK8;RALBP1;PRDX5;CCL21;PLG;CCR2;CCR6;FGF2;HRAS;RIPK2;TACR1;ADM;CXCL10;PROC;MAPK14;ITGA2;RELA;INHBA |
| Regulation of Actin Filament Length | |||||
| 4 | 53 | 6 | 35 | 8.1x10-7 | CXCL12;NCK1;GSN;CAPG |
| Regulation of Actin Polymerization and/or Depolymerization | |||||
| 4 | 53 | 6 | 35 | 8.1x10-7 | CXCL12;NCK1;GSN;CAPG |
| Regulation of Cellular Component Size | |||||
| 4 | 53 | 6 | 35 | 8.1x10-7 | CXCL12;NCK1;GSN;CAPG |
| Behavior | |||||
| 41 | 126 | 22 | 69 | 1.4x10-6 | IL8;CXCL12;CCL20;IL4;IL10;CCL5;IL1RAPL1;CRHBP;LEP;PIK3CB;HPRT1;SOD1;MAPK1;FOSL1; CDH13;PLD1;CXCL11;C5;CXCR4;NPY;DEFA1;PF4;CCR5;TGFB2;CXCL5;CCL27;NF1;MAP2K1;CCR7; CXCR3;PLAUR;KLK8;RALBP1;CCL21;CCR2;CCR6;FGF2;HRAS;TACR1;CXCL10;MAPK14 |
| Actin Polymerization and/or Depolymerization | |||||
| 9 | 62 | 8 | 38 | 3.1x10-6 | CXCL12;NCK1;RAC1;DSTN;ANG;GSN;WIPF1;CAPG;WASL |
| Regulation of Organelle Organization and Biogenesis | |||||
| 11 | 65 | 9 | 39 | 5.2x10-6 | CXCL12;KATNB1;NCK1;MAPRE1;GSN;ARAP1;APC;CAPG;NEXN;NF2;TSC1 |
| Regulation of Cytoskeleton Organization and Biogenesis | |||||
| 11 | 65 | 9 | 39 | 5.2x10-6 | CXCL12;KATNB1;NCK1;MAPRE1;GSN;ARAP1;APC;CAPG;NEXN;NF2;TSC1 |
| Response to Biotic Stimulus | |||||
| 26 | 99 | 16 | 52 | 5.2x10-6 | VCP;TNF;CXCL12;GSK3B;FGR;IL10;CCL5;S100A7;BCL3;HSPB1;CD24;IFNGR1;FOSL1;BCL2;PTPRC; TP53;CXCR4;TLR3;CCL4;IFNAR1;IL12A;VAPB;S100A12;AMFR;LALBA;DMBT1 |
| Nucleus | |||||
| 198 | 318 | 113 | 80 | 1.3x10-5 | SUZ12;GADD45A;BCAS2;VCP;EGFR;HLF;AXIN2;IRS1;GSK3B;FOS;EGR1;KPNA2;BRCA1;CNOT7; SMAD4;CEP290;XPA;MDM2;PARK7;NBN;NCOR2;BRCA2;STK17A;LGALS3;DEK;FOXO3;RPS6KA5; HEXIM1;GZMB;MECP2;CDK1;S100A7;ERG;SUFU;PARP1;SIK1;BCL3;NCOA6;PTGS2;RAD54L; MCM2;PPIA;SPDYA;BIRC3;KDM5B;PTMS;DLGAP5;BCL6;PHB;TP63;CDK6;CRK;MYC;STIP1;NDC80;S100A11;ANG;PTGES3;SOD1;ABL1;BTG1;ACTB;USP4;PPARA;PPIE;FOSL1;FANCA;CYCS;BCL2; METTL3;CHEK1;STAT1;SIN3A;RALY;USF1;CCNA2;MGMT;ASH2L;WRN;GLI3;MALT1;ERCC5;EPC1; CDK9;ESR2;LYN;NFKB2;FOXO1;CDKN1B;TP53;HNRNPU;HTATIP2;APC;PIN1;MED12;PBX3;TBP; G3BP1;GNL3;CDK2;ATXN3;RXRB;FLNA;NOTCH4;MEF2A;CDKN2A;SRF;IGFBP3;USP7;NME1; GATA2;CCNB1;CREB1;MYCN;CHMP1A;CSNK2A1;EZH2;MAPKAPK3;PPM1D;JUN;RARB;NFKBIA; RGS12;XRCC6;PHB2;NPM1;MAML2;HIF1A;NF1;TLE1;TAF1;SMARCA2;SUMO1;SENP2;NAP1L1; NOTCH1;NAP1L3;AXIN1;CFL1;XRCC4;PDZD2;STAT3;CDKN2B;KDM5A;MSH6;CSTF1;CIDEA;CEBPG;PPIG;CDK4;NUP62;CLIC1;MSMB;EED;MRPL40;MLH3;PTGS1;MUTYH;KHDRBS3;LIMK2;NLRP5; RXRA;PA2G4;NOTCH2;DUSP4;ATF4;HDAC1;UBR5;RB1;CDKN2C;RAD50;RAD51;REC8;NF2;SENP1;RANBP1;HDAC2;PSEN2;CDK2AP1;VHL;NCBP2;UTP20;CCS;LSM1;ZBTB22;IRF9;BUB1B;EHD2;EIF6;RAD52;TOB2;ETS2;XRCC5;MAPK14;CASP8AP2;RELA;TRAF4;PTMA |
| Microtubule-Based Process | |||||
| 13 | 67 | 11 | 40 | 2.3x10-5 | CXCL12;KATNB1;NCK1;KPNA2;MAPRE1;GSN;ARAP1;TTK;APC;MAP7;CAPG;NF2;TSC1 |
| Response to Other Organism | |||||
| 20 | 87 | 14 | 46 | 3.5x10-5 | TNF;CXCL12;FGR;IL10;CCL5;S100A7;BCL3;CD24;IFNGR1;FOSL1;BCL2;PTPRC;CXCR4;TLR3;CCL4; IFNAR1;IL12A;S100A12;LALBA;DMBT1 |
| Cell Fraction | |||||
| 76 | 181 | 38 | 82 | 3.8x10-5 | CEACAM1;IL13;VCP;BID;IRS1;TNF;CD55;MDM2;NMB;CD40LG;FBLN5;CRHBP;EEA1;GALK1;LYVE1;AGT;FSHB;EFNB1;IL15;BCAR1;FOLR3;FUT3;LY6D;ATP8B1;STX16;DUSP6;JUP;FUT4;STEAP2;CD59;CYTL1;NTS;HLAB;SPINT2;GP1BA;ANXA2;TP53;PRKCE;WISP2;EDN1;DPP10;APOB;XRCC6; TNFSF13B;WNT5A;SPTAN1;CTTN;ABCB1;DAG1;F2;ACP1;S100A12;CLIC1;FAS;CDH5;PTGS1; ABCA3;SLC1A1;GCLM;LRP12;AMFR;PRKCA;UBR5;CCR2;SOD3;SELP;PRKCZ;CCS;ADM;CD70; WISP1;LRP1;REG3A;PDLIM5;FOLH1;SLC2A1 |
1 Among the total number of genes in the pathway, number of unique genes in our array data
2 Total number of corresponding antibody probes in the assay
3 Among the significant probes, via paired t-test comparing G&C to placebo, the number that have an effect size >0
4 Among the significant probes, via paired t-test comparing G&C to placebo, the number that have an effect size <0
5 GO = Gene Ontology; KEGG = Kyoto Encyclopedia of Genes and Genomes
*All pathways listed were statistically significant with Bonferroni correction (P <3.85x10-4 for KEGG pathway; P<5.18x10-5 for GO pathway)
Of the individual proteins, 508 were statistically significantly different between the two interventions with a Bonferroni correction (p < 1.7 x 10-5). The 100 most significant antibodies (p values 3 x 10-16 to 1 x 10-9) are given in Table 4.
Table 4. Top 100 (of 508) individual protein antibodies significantly different after glucosamine and chondroitin supplementation versus placebo intervention periods (n = 18).
| Gene 1 | Function 2 | Effect size 3 | P value* |
|---|---|---|---|
| CEACAM1 | Cell-cell adhesion | -2.45 | 8.7x10-15 |
| SUZ12 | Proliferation and histone methyltransferase activity [50,51] | 1.09 | 1.7x10-14 |
| THBS4 | Cell-to-cell and cell-to-matrix interactions, extracellular mitogen | -1.88 | 2.1x10-14 |
| GADD45A | Induced in response to DNA damage | -1.37 | 2.8x10-14 |
| ITGA5 | Adhesion and cell-surface mediated signaling | -1.54 | 9.7x10-14 |
| ITGB4 | Adhesion and cell-surface mediated signaling | -1.89 | 1.2 x10-13 |
| CSF3(GCSF)§ | Cytokine involved in hematopoiesis and induction of granulocytes | -3.06 | 2.1 x10-13 |
| PKNOX1 | RNA polymerase II distal enhancer | 1.43 | 2.8 x10-13 |
| IL13§ | Immunoregulatory cytokine involved in inhibition of allergic reaction, particularly in the airways | -6.29 | 3.5 x10-13 |
| C1orf38 | Mediates macrophage inflammatory response | 3.67 | 4.6 x10-13 |
| SON | Splicing co-factor for cell-cycle progression and DNA-repair, involved in differentiation of hematopoietic cells | 1.02 | 6.4 x10-13 |
| MUC3B | Provides protective barrier against infectious agents at mucosal surfaces | 3.83 | 1.3 x10-12 |
| RUNX1 | Subunit of transcription factor that binds to many enhancers and promoters, involved in development of normal hematopoiesis | 3.93 | 1.4 x10-12 |
| IL17D | Cytokine involved in the stimulation of other cytokines, e.g., IL6, IL8, and CSF | -2.27 | 1.6 x10-12 |
| BCAS2 | Component of pre-mRNA splicesome complex | 1.72 | 2.3 x10-12 |
| KCNE3 | Modulates gating kinetics of potassium voltage channel complexes | 1.75 | 3.2 x10-12 |
| CD44 | Cell adhesion and migration, receptor for hyaluronic acid | 1.50 | 3.3 x10-12 |
| VEPH1 | Function unknown | 1.80 | 3.7 x10-12 |
| HBEGF | Normal heart function, smooth muscle cell proliferation, may be involved in macrophage mediated proliferation | -1.47 | 5.2 x10-12 |
| VCP | Putative ATP-binding protein in vesicle transport and fusion, 26S proteasome function and assembly of peroxisomes | -2.10 | 6.8 x10-12 |
| COMP | Structural integrity of cartilage, potent suppressor of apoptosis in chondrocytes | -2.08 | 7.4 x10-12 |
| IL8§ | Chemokine, chemoattractant and potent angiogenic factor | -2.35 | 9.9 x10-12 |
| CAPN3 (NCL1) | Intracellular protease, binds to titin | -2.09 | 1.0 x10-11 |
| GCM2 | Transcription factor regulating parathyroid development | 1.23 | 1.0 x10-11 |
| PKC | Regulation of cell growth and immune responses | -0.89 | 1.3 x10-11 |
| LASP1 | Regulation of actin-based cytoskeletal activities | -1.42 | 1.4 x10-11 |
| SPP1 (Osteopontin) | Attachment of osteoclasts to the mineralized bone matrix; also a cytokine that upregulates expression of interferon-gamma and interleukin-12 | -6.45 | 1.7 x10-11 |
| EFNB3 | Ligand for Eph receptors involved in migration, repulsion and adhesion during neuronal, vascular and epithelial development | -3.24 | 1.9 x10-11 |
| HOXA4 | Transcription factor that may regulate gene expression, morphogenesis and differentiation | 1.88 | 2.3 x10-11 |
| IL1β | Cytokine involved in inflammatory response | -2.65 | 2.3 x10-11 |
| EGFR§ | Cell proliferation | 1.80 | 3.1 x10-11 |
| PRKCQ | Kinase involved in diverse cellular signaling pathways including T-cell activation, proliferation, differentiation and survival | -1.68 | 3.1 x10-11 |
| ENO1 | Multifunctional glycolytic enzyme involved in glycolysis, growth control, hypoxia tolerance and allergic response | -3.37 | 3.2 x10-11 |
| SULF1 | Inhibits signaling by heparin-dependent growth factors, diminishes proliferation and facilitates apoptosis | 2.39 | 3.2 x10-11 |
| MUC6 | Modulates the composition of the protective mucus layer related to acid secretion or presence of bacteria in the lumen | 1.43 | 3.4 x10-11 |
| HDA-1 | Histone deacetylase; regulation of gene expression | 1.35 | 4.3 x10-11 |
| TACSTD2 | Cell surface receptor that transduces calcium signals | 1.20 | 4.9 x10-11 |
| AXIN2 | Inhibitor of Wnt signaling pathway, down-regulates beta-catenin | 1.89 | 5.1 x10-11 |
| IRS1 | Mediates the control of various cellular processes by insulin | -1.37 | 5.1 x10-11 |
| VPS25 | Formation and sorting of endosomal proteins destined for lysosomal degradation | -1.26 | 5.6 x10-11 |
| ANKRD11 | Inhibits ligand-dependent activation of transcription | 1.34 | 5.8 x10-11 |
| DEFA1;A1B | Antibacterial, fungicidal and antiviral activities | -1.01 | 6.5 x10-11 |
| RASGRF2 | Signal coordination of mitogen-activated protein kinase pathways | 1.07 | 8.3 x10-11 |
| ESM1 | Expressed in endothelial cells in lung and kidney, regulated by cytokines | 3.77 | 8.5 x10-11 |
| DSPP | Dentinogenesis | 2.18 | 8.6 x10-11 |
| BAG1 | Anti-apoptotic factor | 1.52 | 9.3 x10-11 |
| NOV | Cell-adhesion, migration, proliferation, differentiation and survival | -1.79 | 9.3 x10-11 |
| LIFR§ | Cytokine | -4.54 | 1.2 x10-10 |
| EXT2 | Heparin sulfate bioysynthesis | 1.16 | 1.2 x10-10 |
| CXCL12(SDF1)§ | Ligand for G-protein coupled receptor involved in inflammation | -1.57 | 1.2 x10-10 |
| CD55/DAF | Regulation of the complement cascade | 1.90 | 1.3 x10-10 |
| LEF1 | Involved in Wnt signaling and enhances T-cell receptor binding | 2.58 | 1.4 x10-10 |
| LIN28B | Suppressor of microRNA biogenesis | 1.40 | 1.4 x10-10 |
| PRLR§ | Cytokine receptor, interacts with prolactin | 2.39 | 1.6 x10-10 |
| MASP1 | Complement activation | -1.99 | 1.7 x10-10 |
| KRT10 | Cytoskeleton composition | -1.56 | 1.8 x10-10 |
| WAS | Signal transducer for actin cytoskeleton | 3.76 | 1.8 x10-10 |
| FSCN1 | Actin-bundling protein | -1.48 | 2.0 x10-10 |
| NAGLU | Degrades heparin sulfate | -1.85 | 2.1 x10-10 |
| CCL20x | Chemotactic factor | 4.00 | 2.5 x10-10 |
| MMP3 | Extracellular matrix degradation | 1.61 | 2.6 x10-10 |
| DLAT | Pyruvate dehydrogenase complex component which catalyzes the conversion of pyruvate to acetyl CoA | -1.88 | 3.0 x10-10 |
| SP1 | Activator or repressor of transcription of a battery of genes related to cell growth, apoptosis, differentiation and immune response | -1.46 | 3.0 x10-10 |
| EREG | Epidermal growth factor family member | -4.04 | 3.0 x10-10 |
| NOLA2 | Ribosome biogenesis and telomere maintenance | 3.92 | 3.1 x10-10 |
| CALR3 | Expressed in testis, may be required for sperm fertility | -1.22 | 3.2 x10-10 |
| IL4§ | Cytokine produced by activated T cells | -1.04 | 3.3 x10-10 |
| ARHGEF17 | Exchange factor for RhoA GTPases | 1.86 | 3.3 x10-10 |
| STMN1 (S15) | Regulation of filament system destabilization | 3.34 | 3.3 x10-10 |
| SPARC (Osteonectin) | Calcification of collagen in bone; also regulates cell shape and growth through interactions with extracellular matrix and cytokines | -1.05 | 3.3 x10-10 |
| FGR | Negative regulator of cell migration and adhesion | 1.60 | 3.5 x10-10 |
| IL6§ | Cytokine with diverse functions in inflammation, maturation of B cells, osteoblast formation, neuronal differentiation, hematopoiesis and energy mobilization in muscle | 1.33 | 3.8 x10-10 |
| CCL5§ | Chemoattractant for monocytes, memory T helper cells and eosinophils | -2.78 | 4.4 x10-10 |
| CSRP1 | Cellular differentiation | -1.11 | 4.9 x10-10 |
| PEPD | Recycling of proline, may be rate limiting for production of collagen | -1.27 | 5.0 x10-10 |
| LCK | Key signaling molecule in selection and maturation of developing T-cells | 1.22 | 5.2 x10-10 |
| WNT3A | Cell-cell signaling during embyogenesis | -2.95 | 5.2 x10-10 |
| DEPDC1 | Transcriptional co-repressor | 2.52 | 5.5 x10-10 |
| KRT18 | Uptake of thrombin-antithrombin complexes by hepatic cells; filament reorganization | -1.56 | 5.5 x10-10 |
| UBC | Protein degradation | -2.00 | 5.6 x10-10 |
| SERPINB5 | Tumor suppressor | 1.27 | 6.1 x10-10 |
| CCSP-2 | Marker for colon cancer | -1.69 | 6.1 x10-10 |
| EGR1 | Activates transcription of genes required for mitogenesis and differentiation | 1.36 | 6.3 x10-10 |
| DCN | Matrix assembly | 1.85 | 6.4 x10-10 |
| TNFRSF9§ | TNF receptor family | -3.59 | 6.9 x10-10 |
| CD142(F3) | Initiates blood coagulation | -1.29 | 7.3 x10-10 |
| NPEPL1 | Aminopeptidase activity | 2.01 | 7.4 x10-10 |
| KPNA2 | Nuclear transport of proteins | 2.01 | 7.7 x10-10 |
| BRCA1 | Tumor suppressor | 2.08 | 7.7 x10-10 |
| IER3 | Anti-apoptotic factor | 2.56 | 7.8 x10-10 |
| PHLDA2 | Tumor Suppressor | -2.73 | 8.2 x10-10 |
| AK1 | Cellular energy homeostasis and adenine nucleotide metabolism | 2.40 | 8.5 x10-10 |
| F13B | Coagulation factor | -3.38 | 8.5 x10-10 |
| GJA1 (pS373) | Gap junction protein | 1.99 | 8.8 x10-10 |
| STX11 | Protein transport | 1.22 | 9.3 x10-10 |
| TLN1/TLN2 | Cytoskeletal protein | -1.64 | 9.3 x10-10 |
| PPP2CA | Negative control of cell-growth and division | -2.08 | 9.4 x10-10 |
| FUT8 | Catalyzes addition of fucose in alpha 1–6 linkages | 2.36 | 1.0 x10-09 |
| ITGA1 | Adhesion and cell-surface mediated signaling | -1.94 | 1.0 x10-09 |
| CNOT7 | Negative regulator of cell proliferation | -1.34 | 1.1 x10-09 |
1Indicates that protein was in the GO (Gene Ontology) or KEGG (Kyoto Encyclopedia of Genes and Genomes) Cytokines pathway
2Information pertaining to function is derived from PubMed Gene and/or UniProtKB unless otherwise noted
3Values >1 indicate greater antibody expression after supplementation with G&C compared to placebo; values <1 indicate lower expression
*All proteins listed were statistically significant with Bonferroni correction (P <1.70x10-5)
Discussion
In this randomized, crossover trial, we found that G&C supplementation statistically significantly lowered mean serum CRP concentrations by 23% compared to placebo. In support of this finding, “Cytokine activity” was the most significant pathway altered between the two interventions in both the GO and KEGG gene-set enrichment analyses, and was lower after G&C. Correspondingly, a number of individual cytokines and other inflammation-related factors were significantly lower in the proteomics panel, including several interleukins and chemokines.
Our results are in agreement with a growing body of in vitro and animal research which suggests that G&C have anti-inflammatory properties. Laboratory studies indicate that G&C inhibit NF-κB, by preventing the degradation of its inhibitory subunit, Iκ-B [10]. Thus, NF-κB is unable to translocate to the nucleus, where this transcription factor activates the expression of a battery of genes involved in inflammatory response and cell proliferation [32]. These anti-inflammatory effects have been corroborated in vivo in animal studies. In rabbits with induced atherosclerosis and chronic arthritis, treatment with G inhibited NF-κB activation and down-regulated COX-2 expression in peripheral blood mononuclear cells, while also decreasing circulating levels of CRP and IL-6 [33]. Further research has shown that mice fed a G-containing diet for 56 days had lower serum IL-6 and TNFα than mice fed a control diet [34]. Two recent studies reported that G&C administration has anti-inflammatory effects in the colon [35]. Bak, et al [13], reported reduced TNFα, interleukin-1β, and NF-κB mRNA expression in colonic mucosa in mice induced with colitis after supplementation of G at 0.10% diet (wet weight) compared to control for four weeks. Finally, in rats with chemically-induced colitis, treatment with G ameliorated symptoms of colitis, while decreasing both systemic and colonic inflammation, as measured by serum concentrations of IL-8 and amyloid P component, and colonic expression of NF-kB, IL-1β, and TNFα, respectively [35]. These studies are notable given our findings of decreased colorectal cancer incidence with use of G&C in the VITAL study [4], and significantly decreased concentrations of CEACAM1 (carcinoembryonic antigen-related cell adhesion molecule 1) in the present study. CEACAM1 is an immunoglobulin involved in cell-to-cell adhesion, and its overexpression is implicated in colorectal cancer [36]. Further, this protein was the most significantly altered protein between the two interventions (effect size-2.45; P = 8.7x10-15).
Despite the compelling results from in vitro and preclinical models, few studies have evaluated the effects of G&C on inflammation in humans. In a nationally representative sample of nearly 10,000 adults included the National Health and Nutrition Examination Study (NHANES), regular use of G&C compared to nonuse was associated with a ~20% lower mean concentrations of hsCRP [14]. In addition, we recently observed G&C use to be associated with 28–36% lower CRP concentrations, [37] and 40–47% lower oxidative stress, as measured by urinary prostaglandin F2α (PGF2α) [38] among a sub-set of 220 individuals in the VITAL cohort [15]. Only two randomized trials of G &/or C on markers of inflammation have been conducted. In a small trial, 36 osteoarthritis patients were given 1500 mg G and 675 mg C for twelve weeks [39]. A significant reduction in serum PGE2 was observed among the G&C treated group, with post-treatment levels similar to that of 25 age-matched healthy controls. The same group conducted a second trial in which 51 rheumatoid arthritis patients were randomized to receive either 1500 mg G or placebo for twelve weeks [40]. While treatment with G reduced serum MMP-3 levels, CRP concentrations remained unchanged. However, exposure was limited to G alone (without C) and patients continued taking regular medications throughout the study. Furthermore, as individuals with rheumatoid arthritis have higher levels of inflammation than the general population, the results of this study may not be generalizable to a healthy population.
Additional inflammation-related pathways that were significantly different between interventions with Bonferroni correction included JAK/STAT signaling, a signaling pathway for a broad range of cytokines and growth factors [41]; intestinal immune network for IgA production, which serves as the first line of defense against microorganisms in the intestinal mucosa [42]; leukocyte transendothelial migration, the movement of leukocytes from blood into tissues for immune surveillance and inflammation [43]; and hematopoietin/interferon cytokine receptor binding, which includes class I cytokines, mainly the interleukins, and growth factors. G-protein coupled receptors are involved in diverse biologic functions, but include roles in growth and regulation of inflammation [44]. As with the cytokine pathway, the majority of the probes in these pathways were less abundant after G&C as compared to placebo. In fact, of the 30 significant pathways, the majority of probes were less abundant in all but two pathways—ubiquitin-mediated proteolysis and proteins contained in the nucleus. Alterations in these other inflammation-related pathways suggest that G&C may have wider-ranging effects on inflammation beyond inhibition of transcription factor NF-kB.
Significant differences in other pathways between the interventions indicate that G&C may play a role in other biologic functions that have not been previously associated with this supplement. For example, pathways related to microtubule function, regulation of actin polymerization, filament length, cytoskeleton organization, and locomotory activity were lower after G&C. Although there is overlap among some genes representing the pathways, these differences suggest that G&C may have effects on cell division and motility, targets of a growing number of chemotherapeutic drugs [45–47]. Lower protein concentrations in pathways related to external and biotic stimulus, and other organisms were likely related to reduction in inflammation-related signaling, particularly given the antibodies contained on our assay. Other pathways that differed between the interventions were based on cellular component or location. The meaning of these differences in unrelated proteins, in terms of health consequences, is unclear.
Among the most significant individual proteins that were differentially abundant between the interventions in the proteomics analysis, several in the top ten were related to cell-adhesion (CEACAM, THBS4, ITGA and B). Other proteins with large effect sizes were IL-13 (down; -6.29), an immunoregulatory cytokine involved in inhibition of allergic reaction, particularly in the airways; osteopontin (down; -6.45), which is involved in the attachment of osteoclasts to the mineralized bone matrix, and is also a cytokine that upregulates the expression of interferon-gamma and IL-12; and C1orf38 (up; 3.67), involved in mediating macrophage inflammatory response. Consistent with the lower CRP concentrations measured in serum, CRP was also down-regulated in plasma on the antibody array (down; -0.5, P = 0.001) but was not significant with correction for multiple testing. As has been observed in many other serum/plasma studies [48,49], some of the proteins with significant changes are normally cytoplasmic or nuclear, so their role as putative serum/plasma biomarkers is unclear. They could indicate sufficient apoptosis and/or necrosis of cells to be detectable and imply that this is reduced with G&C treatment.
This is the first randomized trial to evaluate the effects of G&C on inflammation in healthy adults, and the first to assess potential pathways perturbed by G&C using a broad proteomic screen and gene set enrichment analyses. A major strength of this study is that participants were healthy and free of underlying inflammatory conditions, and the stringent inclusion and exclusion criteria minimized effects of other factors that may affect inflammatory status, (e.g., age, tobacco or medication use, chronic health conditions). Despite the small sample size, many of the intervention effects in the proteomics pathway analyses and for individual proteins were highly significant, exceeding by orders of magnitude, the Bonferroni corrections and the effects observed in multiple previous analyses we have performed in which we compared plasma samples in cancer cases and controls [23,26,27]. Detection of these highly significant effects appears to be due to our use of the randomized, crossover, placebo-controlled design where each participant acted as his or her own control. A strength of the platform is the significant coverage of the proteome including many cytokines (~3,000 proteins). Thus, it is a powerful approach for identifying pathways that change in response to an intervention. Furthermore, it is a powerful discovery method for identifying individual biomarkers that may play an important role in the process being studied. Individual protein results warrant validation by other means such as ELISA or immunoblot since the array uses only a single antibody to bind the protein from plasma and the indicated antibody could bind a protein in plasma that is not the one claimed by the manufacturer (i.e., the specificity of each antibody has not be determined). Finally, given the sample size of the current study, the anti-inflammatory properties of G&C need to be replicated in larger randomized trials and should evaluate sex-specific effects.
In summary, we found that G&C significantly reduced circulating CRP concentrations compared to placebo, and gene set enrichment analyses indicated that cytokine activity and other inflammation-related pathways were significantly decreased. These results are supported by in vitro and animal studies demonstrating anti-inflammatory properties of G&C, as well as human observational data which show an association between G&C use and lower concentrations of circulating CRP. Thus, there is now growing evidence that G&C reduce systemic inflammation in humans. This evidence may also provide a possible biologic mechanism to support prior findings that use of G&C supplements is associated with reduced lung and colon cancer and overall mortality. Future studies in larger samples and other populations are needed to determine the potential utility of G&C as a possible anti-inflammatory agent.
Supporting Information
(DOC)
(XLSX)
(XLS)
(DOC)
Acknowledgments
Glucosamine and chondroitin supplement and placebo were generously donated by Nutramax Laboratories Consumer Care, Inc. (Edgewood, MD). Characterization of trial materials was supported by the Office of Dietary Supplements, NIH (Bethesda, MD). Analysis of PGE-M and F2-isoprostane were performed in the Vanderbilt University Eicosanoid Core Laboratory. Other inflammatory biomarkers were performed at the Biomarker Laboratory, and proteomics assays were carried out in Paul Lampe’s Laboratory at Fred Hutchinson Cancer Research Center; and participant clinic visits for the trial were conducted in the Prevention Center Shared Resource of the Fred Hutchinson Cancer Research Center.
Data Availability
Data from this study may be obtained through the corresponding author, Sandi L. Navarro at the Fred Hutchinson Cancer Research Center
Funding Statement
This work was supported by National Institutes of Health/National Cancer Institute grant NCI P30 CA015704-38. S. L. Navarro was supported by T32 CA09168; E. D. Kantor was supported by the National Cancer Institute, National Institutes of Health (T32CA009001); E. White was supported by a grant from National Cancer Institute and Office of Dietary Supplements (K05-CA154337). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Qato DM, Alexander GC, Conti RM, Johnson M, Schumm P, et al. (2008) Use of prescription and over-the-counter medications and dietary supplements among older adults in the United States. JAMA 300: 2867–2878. 10.1001/jama.2008.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jordan KM, Arden NK, Doherty M, Bannwarth B, Bijlsma JW, et al. (2003) EULAR Recommendations 2003: an evidence based approach to the management of knee osteoarthritis: Report of a Task Force of the Standing Committee for International Clinical Studies Including Therapeutic Trials (ESCISIT). Ann Rheum Dis 62: 1145–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang W, Moskowitz RW, Nuki G, Abramson S, Altman RD, et al. (2008) OARSI recommendations for the management of hip and knee osteoarthritis, Part II: OARSI evidence-based, expert consensus guidelines. Osteoarthritis Cartilage 16: 137–162. 10.1016/j.joca.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 4. Satia JA, Littman A, Slatore CG, Galanko JA, White E (2009) Associations of herbal and specialty supplements with lung and colorectal cancer risk in the VITamins and Lifestyle study. Cancer Epidemiol Biomarkers Prev 18: 1419–1428. 10.1158/1055-9965.EPI-09-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pocobelli G, Kristal AR, Patterson RE, Potter JD, Lampe JW, et al. (2010) Total mortality risk in relation to use of less-common dietary supplements. Am J Clin Nutr 91: 1791–1800. 10.3945/ajcn.2009.28639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bell GA, Kantor ED, Lampe JW, Shen DD, White E (2012) Use of glucosamine and chondroitin in relation to mortality. Eur J Epidemiol 27: 593–603. 10.1007/s10654-012-9714-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bartsch H, Nair J (2006) Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg 391: 499–510. [DOI] [PubMed] [Google Scholar]
- 9. Tudek B, Speina E (2012) Oxidatively damaged DNA and its repair in colon carcinogenesis. Mutat Res 736: 82–92. 10.1016/j.mrfmmm.2012.04.003 [DOI] [PubMed] [Google Scholar]
- 10. Iovu M, Dumais G, du Souich P (2008) Anti-inflammatory activity of chondroitin sulfate. Osteoarthritis Cartilage 16 Suppl 3: S14–18. 10.1016/j.joca.2008.06.008 [DOI] [PubMed] [Google Scholar]
- 11. Largo R, Alvarez-Soria MA, Diez-Ortego I, Calvo E, Sanchez-Pernaute O, et al. (2003) Glucosamine inhibits IL-1beta-induced NFkappaB activation in human osteoarthritic chondrocytes. Osteoarthritis Cartilage 11: 290–298. [DOI] [PubMed] [Google Scholar]
- 12. Chan PS, Caron JP, Orth MW (2006) Short-term gene expression changes in cartilage explants stimulated with interleukin beta plus glucosamine and chondroitin sulfate. J Rheumatol 33: 1329–1340. [PubMed] [Google Scholar]
- 13. Bak YK, Lampe JW, Sung MK (2014) Dietary supplementation of glucosamine sulfate attenuates intestinal inflammation in a mouse model of experimental colitis. J Gastroenterol Hepatol 29: 957–963. 10.1111/jgh.12485 [DOI] [PubMed] [Google Scholar]
- 14. Kantor ED, Lampe JW, Vaughan TL, Peters U, Rehm CD, et al. (2012) Association between use of specialty dietary supplements and C-reactive protein concentrations. Am J Epidemiol 176: 1002–1013. 10.1093/aje/kws186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kantor ED, Ulrich CM, Owen RW, Schmezer P, Neuhouser ML, et al. (2013) Specialty supplement use and biologic measures of oxidative stress and DNA damage. Cancer Epidemiol Biomarkers Prev 22: 2312–2322. 10.1158/1055-9965.EPI-13-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herder C, Schneitler S, Rathmann W, Haastert B, Schneitler H, et al. (2007) Low-grade inflammation, obesity, and insulin resistance in adolescents. J Clin Endocrinol Metab 92: 4569–4574. [DOI] [PubMed] [Google Scholar]
- 17. Fenkci S, Rota S, Sabir N, Sermez Y, Guclu A, et al. (2006) Relationship of serum interleukin-6 and tumor necrosis factor alpha levels with abdominal fat distribution evaluated by ultrasonography in overweight or obese postmenopausal women. J Investig Med 54: 455–460. [DOI] [PubMed] [Google Scholar]
- 18. Uwe S (2008) Anti-inflammatory interventions of NF-kappaB signaling: potential applications and risks. Biochem Pharmacol 75: 1567–1579. [DOI] [PubMed] [Google Scholar]
- 19. Aziz N, Fahey JL, Detels R, Butch AW (2003) Analytical performance of a highly sensitive C-reactive protein-based immunoassay and the effects of laboratory variables on levels of protein in blood. Clin Diagn Lab Immunol 10: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Navarro SL, Brasky TM, Schwarz Y, Song X, Wang CY, et al. (2012) Reliability of serum biomarkers of inflammation from repeated measures in healthy individuals. Cancer Epidemiol Biomarkers Prev 21: 1167–1170. 10.1158/1055-9965.EPI-12-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fitzgerald DW, Bezak K, Ocheretina O, Riviere C, Wright TC, et al. (2012) The effect of HIV and HPV coinfection on cervical COX-2 expression and systemic prostaglandin E2 levels. Cancer Prev Res (Phila) 5: 34–40. 10.1158/1940-6207.CAPR-11-0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Milne GL, Gao B, Terry ES, Zackert WE, Sanchez SC (2013) Measurement of F2- isoprostanes and isofurans using gas chromatography-mass spectrometry. Free Radic Biol Med 59: 36–44. 10.1016/j.freeradbiomed.2012.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loch CM, Ramirez AB, Liu Y, Sather CL, Delrow JJ, et al. (2007) Use of high density antibody arrays to validate and discover cancer serum biomarkers. Mol Oncol 1: 313–320. 10.1016/j.molonc.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rho JH, Lampe PD (2013) High-throughput screening for native autoantigen-autoantibody complexes using antibody microarrays. J Proteome Res 12: 2311–2320. 10.1021/pr4001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramirez AB, Loch CM, Zhang Y, Liu Y, Wang X, et al. (2010) Use of a single-chain antibody library for ovarian cancer biomarker discovery. Mol Cell Proteomics 9: 1449–1460. 10.1074/mcp.M900496-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li CI, Mirus JE, Zhang Y, Ramirez AB, Ladd JJ, et al. (2012) Discovery and preliminary confirmation of novel early detection biomarkers for triple-negative breast cancer using preclinical plasma samples from the Women’s Health Initiative observational study. Breast Cancer Res Treat 135: 611–618. 10.1007/s10549-012-2204-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramirez AB, Lampe PD (2010) Discovery and validation of ovarian cancer biomarkers utilizing high density antibody microarrays. Cancer Biomark 8: 293–307. 10.3233/CBM-2011-0215 [DOI] [PubMed] [Google Scholar]
- 28. Smyth GK (2005) Limma: linear models for microarray data In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber I.W, editor. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; pp. 397–420. [Google Scholar]
- 29. Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- 30. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, et al. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pahl HL (1999) Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18: 6853–6866. [DOI] [PubMed] [Google Scholar]
- 33. Largo R, Martinez-Calatrava MJ, Sanchez-Pernaute O, Marcos ME, Moreno-Rubio J, et al. (2009) Effect of a high dose of glucosamine on systemic and tissue inflammation in an experimental model of atherosclerosis aggravated by chronic arthritis. Am J Physiol Heart Circ Physiol 297: H268–276. 10.1152/ajpheart.00142.2009 [DOI] [PubMed] [Google Scholar]
- 34. Azuma K, Osaki T, Wakuda T, Tsuka T, Imagawa T, et al. (2012) Suppressive effects of N-acetyl-D-glucosamine on rheumatoid arthritis mouse models. Inflammation 35: 1462–1465. 10.1007/s10753-012-9459-0 [DOI] [PubMed] [Google Scholar]
- 35. Yomogida S, Kojima Y, Tsutsumi-Ishii Y, Hua J, Sakamoto K, et al. (2008) Glucosamine, a naturally occurring amino monosaccharide, suppresses dextran sulfate sodium-induced colitis in rats. Int J Mol Med 22: 317–323. [PubMed] [Google Scholar]
- 36. Beauchemin N, Arabzadeh A (2013) Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev 32: 643–671. 10.1007/s10555-013-9444-6 [DOI] [PubMed] [Google Scholar]
- 37. Kantor ED, Lampe JW, Navarro SL, Song X, Milne GL, et al. (2014) Associations between glucosamine and chondroitin supplement use and biomarkers of systemic inflammation. J Altern Complement Med 20: 479–485. 10.1089/acm.2013.0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yin H, Gao L, Tai HH, Murphey LJ, Porter NA, et al. (2007) Urinary prostaglandin F2alpha is generated from the isoprostane pathway and not the cyclooxygenase in humans. J Biol Chem 282: 329–336. [DOI] [PubMed] [Google Scholar]
- 39. Nakamura H, Nishioka K (2002) Effects of glucosamine/chondroitin supplement on osteoarthritis: Involvement of PGE2 and YKL-40. J Rheumatism Joint Surgery 21: 175–184. [Google Scholar]
- 40. Nakamura H, Masuko K, Yudoh K, Kato T, Kamada T, et al. (2007) Effects of glucosamine administration on patients with rheumatoid arthritis. Rheumatol Int 27: 213–218. [DOI] [PubMed] [Google Scholar]
- 41. Rawlings JS, Rosler KM, Harrison DA (2004) The JAK/STAT signaling pathway. J Cell Sci 117: 1281–1283. [DOI] [PubMed] [Google Scholar]
- 42. Cerutti A, Rescigno M (2008) The biology of intestinal immunoglobulin A responses. Immunity 28: 740–750. 10.1016/j.immuni.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muller WA (2011) Mechanisms of leukocyte transendothelial migration. Annu Rev Pathol 6: 323–344. 10.1146/annurev-pathol-011110-130224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kroeze WK, Sheffler DJ, Roth BL (2003) G-protein-coupled receptors at a glance. J Cell Sci 116: 4867–4869. [DOI] [PubMed] [Google Scholar]
- 45. Mukhtar E, Adhami VM, Mukhtar H (2014) Targeting microtubules by natural agents for cancer therapy. Mol Cancer Ther 13: 275–284. 10.1158/1535-7163.MCT-13-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alberti C (2009) Cytoskeleton structure and dynamic behaviour: quick excursus from basic molecular mechanisms to some implications in cancer chemotherapy. Eur Rev Med Pharmacol Sci 13: 13–21. [PubMed] [Google Scholar]
- 47. Jordan MA, Wilson L (1998) Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr Opin Cell Biol 10: 123–130. [DOI] [PubMed] [Google Scholar]
- 48. Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, et al. (2004) The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol Cell Proteomics 3: 311–326. [DOI] [PubMed] [Google Scholar]
- 49. Zhang Q, Menon R, Deutsch EW, Pitteri SJ, Faca VM, et al. (2008) A mouse plasma peptide atlas as a resource for disease proteomics. Genome Biol 9: R93 10.1186/gb-2008-9-6-r93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao R, Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15: 57–67. [DOI] [PubMed] [Google Scholar]
- 51. Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(XLSX)
(XLS)
(DOC)
Data Availability Statement
Data from this study may be obtained through the corresponding author, Sandi L. Navarro at the Fred Hutchinson Cancer Research Center