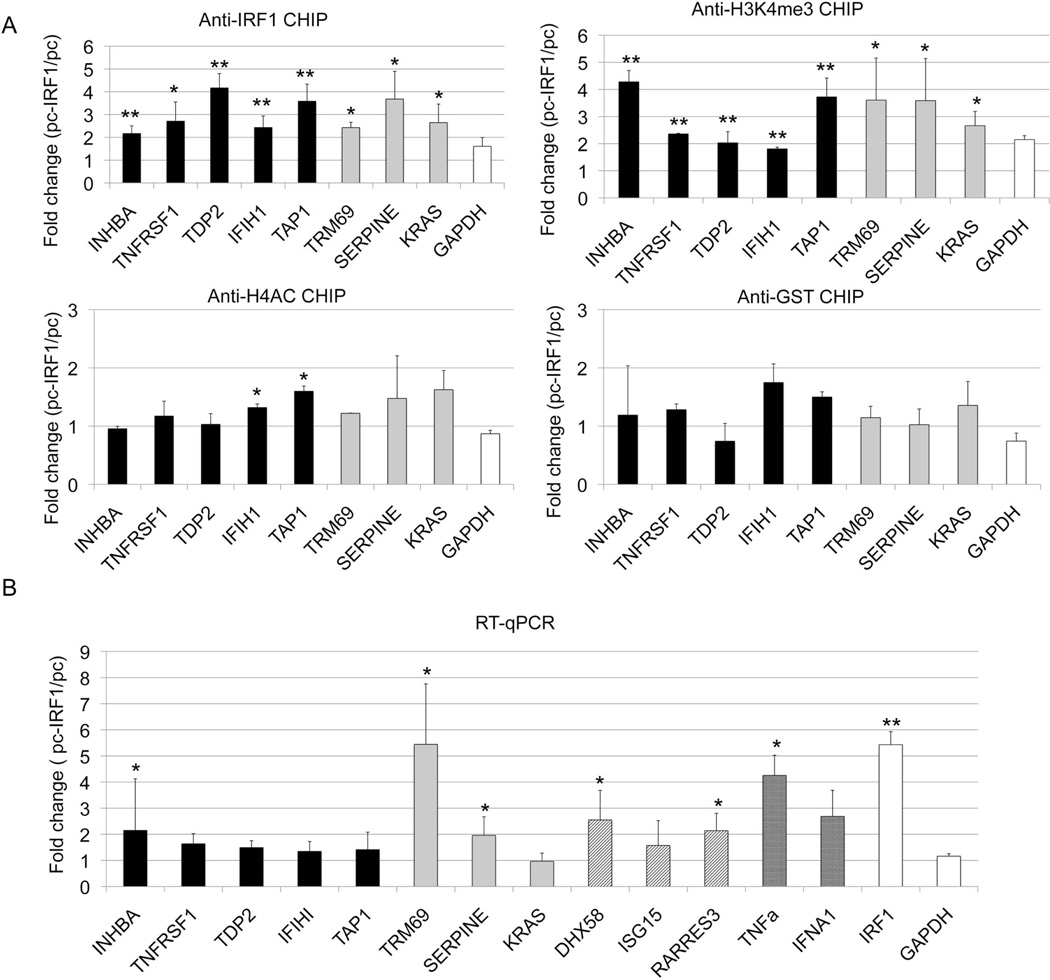
Figure 4.
The effect of IRF1 overexpression on histone modifications. D54MG cells were stably transfected by IRF1 cDNA over-expression vector (pc-IRF1) or control empty vector (pc). A) The cells were immunoprecipitated with antibody to IRF1, H3K4me3, H4ac or GST (a non-binding control antibody). The effect of overexpression was defined. The black bars represent the target genes with both increased IRF1 binding and mRNA expression in SLE patients. The grey bars represent potential IRF1 targets. GAPDH is a control gene, unaltered in SLE. B) RNA was harvested from transfected cells and cDNA was quantitated using qRT-PCR. The black bars represent targets with both increased IRF1 binding and mRNA in SLE (INHBA, TNFRSF14, TAP1, TDP2). The grey bars signify genes with potential IRF1 binding sites (TRIM69, SERPINE1, KRAS). The diagonal lines signify genes with very high IRF1 peaks (DHX58, ISG15, RARRES3, IFIH1). The cross hatched bars represent cytokine genes known to be overexpressed in SLE. IRF1 and GAPDH are shown as controls. Error bars indicate standard error. N=3. Student's t-test was used to determine statistical significance (* indicates p<0.05, ** indicates p<0.005).