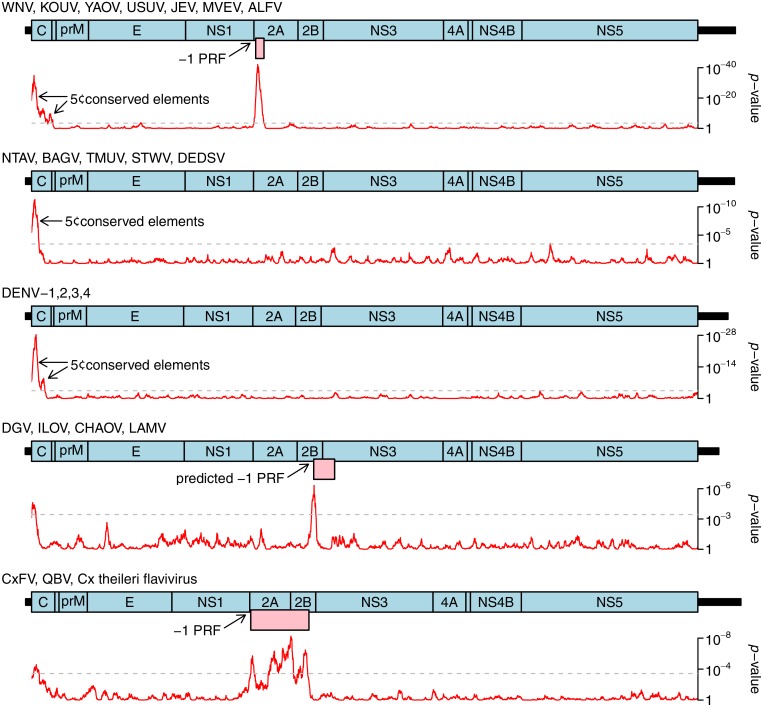
Fig 4. Synonymous site conservation analysis for selected flavivirus clades.
Alignments of 249 JEV serogroup, 49 NTAV/TMUV clade, 89 DENV, 6 DGV/LAMV clade and 29 CxFV/QBV clade polyprotein ORF sequences were analyzed for synonymous site variability as decribed previously (Firth et al., 2011 PMID 21525127). The accession numbers of all sequences used in the analysis are available on request. Red lines indicate the probability (p-value) of obtaining not more than the observed number of synonymous substitutions, in a 25-codon sliding window, under a null model of neutral evolution at synonymous sites. Dashed grey lines indicate an approximate 5% false positive threshold after correcting for multiple tests (i.e. ~136 x 25-codon windows in the ~3400-codon polyprotein ORF). Statistically significant peaks in synonymous site conservation are indicative of overlapping functional elements, either coding or non-coding. Genome maps are shown for each clade. UTR lengths may be uncertain for less well-studied clades. Known and predicted overlapping ORFs accessed via-1 PRF are shown in pink. The predicted overlapping ORF in the DGV/LAMV clade is much shorter in DGV than in other members of the clade; the long form of the ORF is indicated. Note that p-values can not be directly compared between different clades because the statistical significance (i.e. p-value) of observed reductions in synonymous site variabilty depends on the diversity of the specific sequence alignment being analyzed.