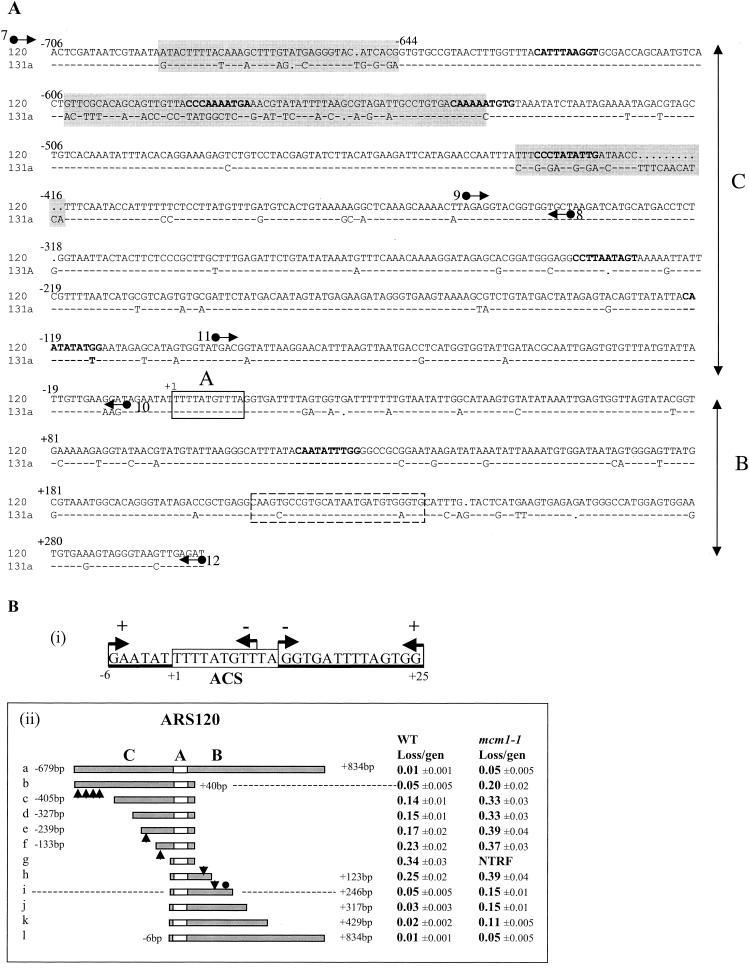
FIG. 3.
Dissection of ARS120. (A) Alignment of ARS120 and ARS131a DNA sequences. Domains C, A, and B are indicated. Nonconserved sequences clustered in the C domain are highlighted. ACS is boxed in solid lines. The Abf1 binding site is boxed in dashed lines. MCEs determined by the weighted matrix score (30, 47) are in bold type. Arrows represent primers used for sequence analysis in Fig. 4 and to generate DNA fragments by PCR for EMSA. Primers 7 and 8 were used to amplify fragment I by PCR. Primers 9 and 10 were used to amplify fragment II. Primers 11 and 12 were used to amplify fragment III. (B) Identification of the essential and important elements of ARS120. (i) Summary of HFT results of 5′- or 3′-end deletions generated by Bal31 digestion of the SacII/StuI fragment of ARS120. Endpoints of 5′ and 3′ deletions are represented by arrows pointing right and left, respectively. A plus sign indicates HFT activity, and a minus sign indicates no HFT activity. The boxed sequence is the ACS. (ii) Stability of minichromosomes containing the deletion fragments of ARS120 in the wild-type (WT) and mcm1-1 mutant strains. Endpoints of each deletion are indicated. Arrowheads represent locations of MCEs identified by the weighted matrix program. The filled circle represents the Abf1 binding site. The empty box represents the ACS. Domains C, B, and A are indicated. gen, generation; NTRF, no transformation.