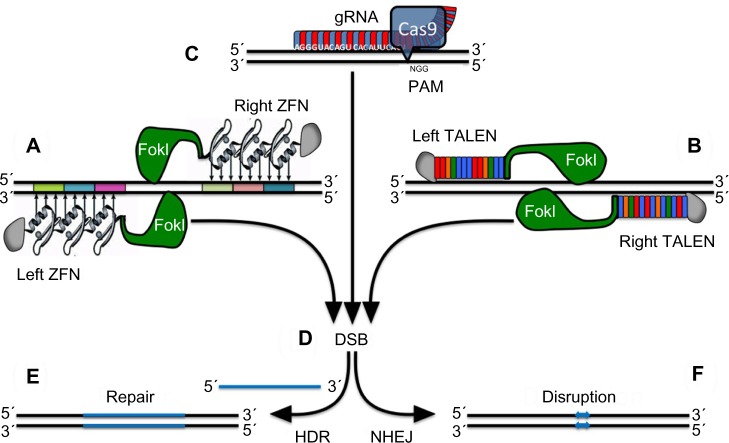
Figure 6.
Alternative strategies for targeted gene repair.
Notes: Schematic representation of (A) ZFNs, (B) TALENs, and (C) a monomeric clustered regularly interspaced short palindromic repeat associated with Cas9 nuclease bound to double-stranded target DNA. (D) The FokI nuclease introduces a nick in the spacer region of the target DNA, necessitating a dimer of two modules to introduce a DSB in the target, while monomeric Cas9 suffices to introduce a DSB preceding its PAM site. Depending on the intended application, the DSB may then be repaired (E) by the template-dependent HDR mechanism (eg, for the repair of a causative mutation) or (F) by error-prone NHEJ (eg, for the disruption of γ-globin repressors or their binding sites). The HDR approach has already been applied by Zou et al158 and Ma et al168 to correct the β-globin gene in induced pluripotent stem cells from sickle cell anemia and β-thalassemia patients, respectively.
Abbreviations: DSB, double-strand break; gRNA, guide RNA; HDR, homology-directed repair; NHEJ, nonhomologous end joining; PAM, protospacer adjacent motif; TALEN, transcription activator-like effector nuclease; ZFN, zinc finger nuclease.