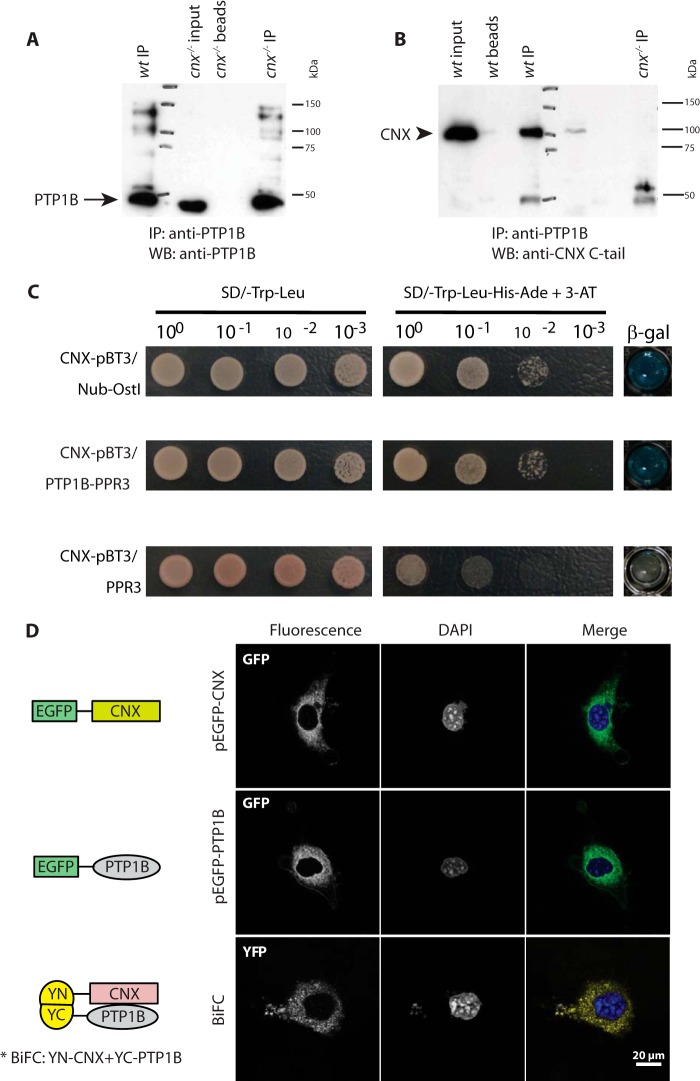
FIGURE 4.
Calnexin and PTP1B form complexes. A, immunoprecipitation was carried out on wild-type (wt) and calnexin-deficient (cnx−/−) fibroblasts with anti-PTP1B antibodies followed by Western blot analysis with anti-PTP1B antibodies. The higher molecular weight banding pattern observed is consistent with SUMO-modification of PTP1B. The arrow indicates the location of the PTP1B protein band. The data represents more than three biological replicates. IP, immunoprecipitation; WB, Western blot. B, immunoprecipitation was carried out with anti-PTP1B antibodies followed by Western blot analysis with anti-calnexin C-tail antibodies. The arrowhead indicates the location of calnexin. The data represent more than three biological replicates. C, DUAL membrane yeast-2-hybrid interaction method was used to determine if PTP1B and CNX interact in situ. Full-length calnexin and PTP1B were cloned into the pBT3-SUC and pPR3 vector, respectively, and co-transformed into the yeast strain NYM51. A positive interaction was determined by β-galactosidase activity. Co-transformation of calnexin and PTP1B demonstrated comparable growth to the CNX-pBT3/Nub-Ostl positive control as well as positive β-galactosidase activity indicating PTP1B and calnexin interact. No β-galactosidase (β-gal) activity was observed with the negative control CNX-pBT3/PPR3. The data represent more than three biological replicates. D, GFP-tagged calnexin (upper panel) and PTP1B (middle panel) are localized to the ER-like network. BiFC analysis of binding between calnexin and PTP1B in NIH3T3 cells (bottom panel). Calnexin was expressed as fusion protein with the C-terminal fragment of YFP (YN-CNX), whereas PTP1B was expressed as a fusion with the C-terminal portion of YFP (YC-PTP1B). Schematic diagrams of each of the constructs are shown in the figure. The data represent more than three biological replicates.