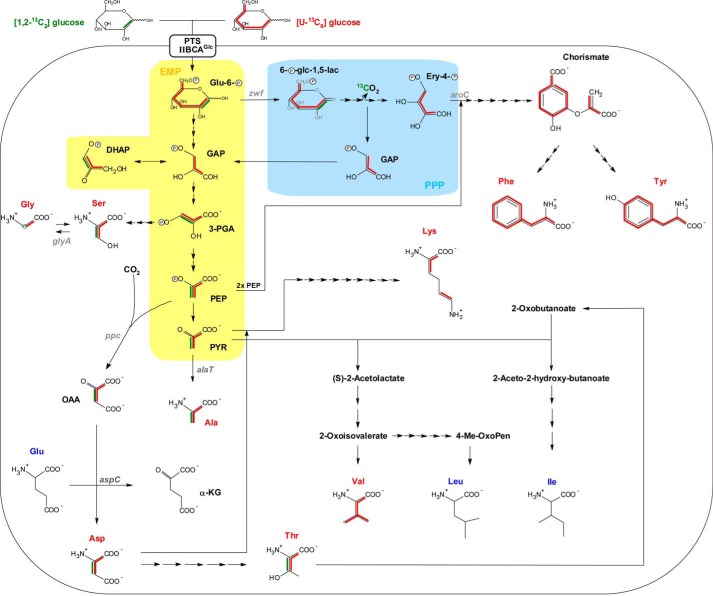
FIGURE 3.
Reconstruction of the metabolic pathways utilized for de novo syntheses of amino acids as derived from isotopologue profiling with [U-13C6]glucose and [1,2-13C2]glucose. 13C patterns of amino acid isotopologues are indicated by red and green lines, respectively, as deduced from labeling experiments performed with [U-13C6]glucose and [1,2-13C2]glucose. [1,2-13C2]Glucose-derived isotopologue patterns are shown for Ala, Asp, Thr, biosynthesis pathways, and the formation of Gly from Ser only. The three-letter abbreviation of de novo synthesized amino acids is highlighted in red, whereas that of unlabeled amino acids remains dark blue. Simplified metabolic reactions being part of the EMP pathway and PPP are highlighted by yellow and light blue shading, respectively. Note that in the case of [1,2-13C2]glucose catabolization via PPP, the formation of unlabeled GAP molecules is shown solely. Full metabolic fluxes refer to the Kyoto Encyclopedia of Genes and Genomes database, and arrows indicate the number of metabolic steps needed for biosynthesis of amino acids. Corresponding gene annotations are given in Table S9. Glu-6-P, glucose-6-phosphate; 6-P-glc-1,5-lac, 6-phospho-d-glucono-1,5-lactone; Ery-4-P, erythrose-4-phosphate; DHAP, dihydroxy-acetone-phosphate; PYR, pyruvate; α-KG, α-ketoglutarate.