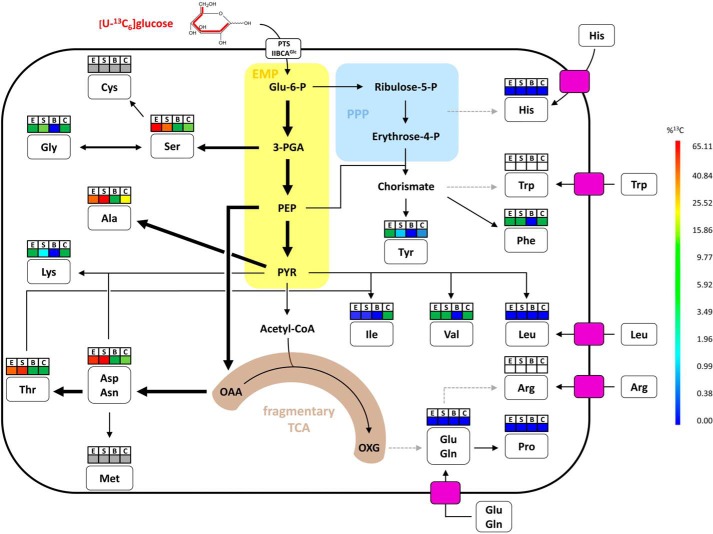
FIGURE 6.
Simplified metabolic model for amino acid biosynthesis pathways of S. suis in vitro and ex vivo. This model is based on the information obtained from isotopologue profiling experiments, amino acid auxotrophy experiments, and S. suis P1/7 genome annotation. Relative metabolic fluxes are shown for isotopologue experiments conducted for S. suis grown in CDM to the exponential (E) growth phase, in CDM to the stationary (S) growth phase, in porcine blood (B), and in porcine CSF (C). Higher relative metabolic fluxes are displayed by thick arrows. Dotted gray arrows indicate that genes required for the biosynthesis of the amino acid are absent in the S. suis P1/7 genome. The colored boxes above amino acids correspond to the mean overall 13C enrichments described for Figs. 2A and 4A. Gray and white boxes stand for amino acids that could not be measured by our protocol but were found to be nonessential and essential, respectively, for growth of S. suis in CDM in vitro. Furthermore, the necessity for uptake of essential amino acids by streptococcal transporters is indicated by pink rectangles. Glu-6-P, glucose-6-phosphate; Acetyl-CoA, acetyl coenzyme A; OXG, oxoglutarate (α-ketoglutarate).