Background: tRNA methyltransferases specifically recognize substrate tRNAs.
Results: To clarify the tRNA recognition mechanism of TrmI, three tRNA species and 45 variants were analyzed in vitro and in vivo.
Conclusion: TrmI recognizes the aminoacyl stem, variable region, C56, purine 57, A58, and U60 in the T-loop of tRNA.
Significance: Our in vitro experimental results explain the regulation of in vivo methylation levels in tRNAs.
Keywords: RNA Methylation, RNA Methyltransferase, RNA Modification, Thermophile, Transfer RNA (tRNA)
Abstract
TrmI generates N1-methyladenosine at position 58 (m1A58) in tRNA. The Thermus thermophilus tRNAPhe transcript was methylated efficiently by T. thermophilus TrmI, whereas the yeast tRNAPhe transcript was poorly methylated. Fourteen chimeric tRNA transcripts derived from these two tRNAs revealed that TrmI recognized the combination of aminoacyl stem, variable region, and T-loop. This was confirmed by 10 deletion tRNA variants: TrmI methylated transcripts containing the aminoacyl stem, variable region, and T-arm. The requirement for the T-stem itself was confirmed by disrupting the T-stem. Disrupting the interaction between T- and D-arms accelerated the methylation, suggesting that this disruption is included in part of the reaction. Experiments with 17 point mutant transcripts elucidated the positive sequence determinants C56, purine 57, A58, and U60. Replacing A58 with inosine and 2-aminopurine completely abrogated methylation, demonstrating that the 6-amino group in A58 is recognized by TrmI. T. thermophilus tRNAGGUThrGGUThr contains C60 instead of U60. The tRNAGGUThr transcript was poorly methylated by TrmI, and replacing C60 with U increased the methylation, consistent with the point mutation experiments. A gel shift assay revealed that tRNAGGUThr had a low affinity for TrmI than tRNAPhe. Furthermore, analysis of tRNAGGUThr purified from the trmI gene disruptant strain revealed that the other modifications in tRNA accelerated the formation of m1A58 by TrmI. Moreover, nucleoside analysis of tRNAGGUThr from the wild-type strain indicated that less than 50% of tRNAGGUThr contained m1A58. Thus, the results from the in vitro experiments were confirmed by the in vivo methylation patterns.
Introduction
To date, more than 90 modified nucleosides have been found in tRNA (1). The majority of these modified nucleosides in tRNA are methylated nucleosides whose biosynthetic pathways include one or multiple methylation steps. These methylated nucleosides in tRNA play important roles in protein synthesis (2). Furthermore, recent studies have elucidated that the modified nucleosides in tRNA are involved in higher biological phenomena such as the RNA quality control system (3, 4), regulation of the subcellular localization of RNA (5), infection (6, 7), and immune responses (8, 9).
1-Methyladenosine (m1A)2 is one of the methylated nucleosides that is found in tRNAs from all three domains of life. This modification has been found at positions 9, 22, 57, and 58 in tRNAs (10). Given that tRNAs from Escherichia coli do not contain the m1A modification at any position, the enzymatic activities involved in generating this modification were identified using E. coli tRNAs as substrates in crude cell extracts from various living organisms, for example Neurospora crassa (11), hamster (12), and HeLa cells (13). Although enzymatic activities involved in generating the m1A modification were detected in various living organisms, it was very difficult to purify the enzymes. Several purification trials were reported from rat liver (14, 15), plant (16), and slime mold (17). However, the genes encoding the enzymes were not identified for a long time.
At the end of the twentieth century, it was reported that the trm6 and trm61 genes (classical names, gcd10 and gcd14) encode tRNA (m1A58) methyltransferase in Saccharomyces cerevisiae (18, 19). Thus, the eukaryotic methyltransferase for m1A58 is a heterodimeric protein. Later, genes for the eubacterial (20) and archaeal (21) enzymes were identified as trmI. Eubacterial TrmI modifies only A58, whereas archaeal TrmI modifies both A57 and A58 (21, 22). Consequently, to distinguish the eubacterial and archaeal enzymes, we abbreviate archaeal TrmI as aTrmI here. TrmI and aTrmI are homotetrameric enzymes. Comparison of the crystal structures of TrmI (23–25) and aTrmI (26) revealed that the tetrameric structure of TrmI is reinforced by salt bridges between the subunits, whereas that of a TrmI from Pyrococcus abyssi is maintained by intersubunit disulfide bonds (27). Thus, eukaryotic Trm6-Trm61, eubacterial TrmI, and aTrmI have different protein architectures even though these enzymes are responsible for the same m1A58 modification in tRNA. In addition, recently, the enzymes responsible for m1A22 in eubacterial tRNA, m1A9 in human mitochondrial tRNA, m1G9 and m1A9 in archaeal tRNA, and m1A58 in mitochondrial tRNA were identified as TrmK (28), a subcomplex of mitochondrial RNase P (29), an archaeal Trm10 homolog (30), and Trm61B (31), respectively.
As mentioned above, the enzymatic activities involved in generating the m1A58 modification were detected using E. coli tRNA in crude cell extracts or with partially purified enzymes. In these early studies, the substrate tRNA specificities of these enzymes were discussed without discriminating the sources of enzymes or the tRNA (14, 15). However, recent studies have showed that eukaryotic Trm6-Trm61, eubacterial TrmI, and aTrmI have different protein architectures. In the current study, we focused on the mechanism by which TrmI from T. thermophilus, an extreme thermophilic eubacterium, recognizes substrate tRNA. Given that all tRNAs reported from T. thermophilus commonly contain the m1A58 modification (32–35), it was thought that eubacterial TrmI has no specificity for particular tRNAs. In this study, we show that TrmI from T. thermophilus does in fact prefer to act on specific tRNAs.
EXPERIMENTAL PROCEDURES
Materials
[methyl-14C]-S-Adenosyl-l-methionine (AdoMet) (1.95 GBq/mmol) and [methyl-3H]AdoMet (2.89 TBq/mmol) were purchased from ICN. Non-radioisotope-labeled AdoMet and standard modified nucleoside (m1A) were obtained from Sigma. HiTrap Heparin HP, Superdex 75 prep grade (pg), and Q-Sepharose Fast Flow were purchased from GE Healthcare. DNA oligomers were bought from Invitrogen. All other chemical reagents were of analytical grade.
Construction of Recombinant TrmI Expression System
Briefly, the trmI gene from T. thermophilus HB8 was amplified by the polymerase chain reaction using the following primers: TrmI forward primer, 5′-GGG CAT ATG GCG TGG CCG GGA CCG CTA CTC-3′; TrmI reverse primer, 5′-GGG GGA TCC TTA GGA GGC CTT CCA TCG CCT AAG-3′. Underlined nucleotides correspond to restriction enzyme sites (NdeI and BamHI, respectively). The amplified DNA was inserted between the NdeI and BamHI sites of the pET30a plasmid vector (Novagen). Recombinant TrmI was expressed at 37 °C for 4 h by induction with isopropyl β-d-thiogalactopyranoside using E. coli BL21(DE3) Rosetta 2 (Novagen).
Purification of Recombinant TrmI Protein
Wet cells (10 g) were suspended in 50 ml of buffer A (50 mm Tris-HCl (pH 7.6), 5 mm MgCl2, 100 mm KCl) and then disrupted with an Ultrasonic Disruptor Model UD-200 (Tomy, Japan). The cell debris was removed by centrifugation at 8000 × g at 4 °C for 20 min, and then the supernatant fraction was heated at 70 °C for 30 min. The denatured proteins were removed by centrifugation at 24,000 × g at 4 °C for 20 min. The supernatant fraction was loaded onto a HiTrap Heparin HP column, and the proteins were eluted with a linear gradient of 100–1000 mm KCl in buffer A. The eluted fractions of TrmI were subjected to 15% SDS-polyacrylamide gel electrophoresis (PAGE), collected, and concentrated with Centriprep YM-10 centrifugal filter devices (Millipore). The resultant sample was loaded onto a Superdex 75 prep grade column equilibrated with buffer A that contained 200 mm KCl. The eluted fractions of TrmI were combined and concentrated. Glycerol was added to a final concentration of 50%, and the protein was stored at −30 °C.
Preparation of Variant tRNA Transcripts
RNA transcripts were prepared by transcription with T7 RNA polymerase. The transcripts were purified by chromatography through a Q-Sepharose Fast Flow column and 10% PAGE (7 m urea). Transcripts 15, 16, 17, and 18 were chemically synthesized by Sigma-Aldrich. The tRNA transcript that contained inosine (I) or 2-aminopurine (2-AP) at position 58 was prepared as follows. The 5′-half that corresponded to G1–G36 of T. thermophilus tRNAPhe was synthesized with T7 RNA polymerase. The 3′-half that contained I (5′-AAU CGC AGU GUC GGC GGU UCG IUU CCG CUC CUC GGC ACC A-3′) and corresponded to A37–A76 was synthesized by Gene Design Co. Ltd. (Japan). The 3′-half that contained 2-AP (5′-AAU CGC AGU GUC GGC GGU UCG 2-APUU CCG CUC CUC GGC ACC A-3′) was synthesized by Sigma-Aldrich. The 5′-end of the 3′-half (1.0 A260 unit) was phosphorylated at 37 °C for 1 h with 10 units of T4 polynucleotide kinase (Takara) and 1 mm ATP. The efficiency of 5′-phosphorylation was checked by 10% PAGE (7 m urea), and the phosphorylated fragment was recovered by phenol-chloroform extraction and ethanol precipitation. The 5′- and 3′-halves (0.75 A260 unit each) were then annealed in 50 μl of buffer B (20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 100 mm NaCl) by cooling from 90 to 40 °C for 50 min. After annealing, 250 units of T4 RNA ligase (Takara), bovine serum albumin (final concentration, 0.01%), and MgCl2 (final concentration, 10 mm) were added to the sample. The sample was incubated at 16 °C for 12 h. The ligated sample was purified by phenol-chloroform extraction and 10% PAGE (7 m urea).
Measurement of TrmI Activity
Incorporation of [14C]methyl group from AdoMet into the T. thermophilus tRNAPhe transcript was used to assay TrmI activity. We used a gel assay described previously to visualize methylated RNAs (36). Briefly, 10 μl of buffer A that contained 0.1 μm TrmI, 20 μm transcript, and 37 μm [methyl-14C]AdoMet were incubated at 55 °C for 5 min and then loaded onto a 10% polyacrylamide gel (7 m urea). To obtain the kinetic parameters of TrmI for each tRNA, the concentrations of TrmI and [methyl-3H]AdoMet (diluted with non-radioisotope-labeled AdoMet) were fixed at 0.1 and 405 μm, respectively. The reaction was basically performed at 55 °C for 5 min, but the incubation time varied from 1 to 15 min depending on the methyl group acceptance activity of each individual transcript. The transcript concentrations in the typical assay were 0, 10, 30, 50, 100, 200, 300, and 500 nm. In the case of tRNAGGUThr variants, [methyl-3H]AdoMet was used for measurements of methyl group acceptance activities in the all experiments because the methyl group acceptance activities of these variants were very low.
Gel Mobility Shift Assay
A 3% agarose gel (Agarose S, Takara) and Tris-acetate-Mg-EDTA buffer (40 mm Tris acetate (pH 8.0), 5 mm MgCl2, 1 mm EDTA) was used for the electrophoresis. tRNA at a concentration of 6.8 μm and TrmI (0, 0.8, 1.3, 2.1, 3.5, 7.7, 11.5, or 19.1 μm) were mixed in 10 μl of buffer (50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 100 mm KCl, 0.1 mm sinefungin, 18% glycerol). Sinefungin (Sigma) is an analog inhibitor for AdoMet-dependent methyltransferases. The samples were incubated at 60 °C for 30 min and then cooled on ice for 10 min. The electrophoresis was performed at 4 °C. The gel was stained with ethidium bromide.
T. thermophilus Wild-type and trmI Gene Disruptant Strains
The culture source of T. thermophilus HB8 was a kind gift from Dr. Tairo Oshima (Tokyo University of Pharmacy and Life Sciences). The cells were grown in rich medium (0.8% polypeptone, 0.4% yeast extract, 0.2% NaCl (pH 7.5 adjusted with NaOH)). The medium was supplemented with 0.35 mm CaCl2 and 0.17 mm MgCl2 after autoclaving. To make plates, gellan gum (Wako Pure Chemicals) was added to the medium (final concentration, 1.5%). The construction of the trmI gene disruptant strain was reported previously (37): the gene was disrupted by replacement with the highly thermostable kanamycin nucleotidyltransferase gene (38, 39).
Purification of Native tRNAPhe and tRNAGGUThr by Solid-phase DNA Probe
Native tRNAPhe was purified in accordance with our previous report (35, 39, 40). To purify tRNAGGUThr, a 3′-biotinylated DNA oligomer (5′-ACC AAG GGT GTG CTC TAC CTG CT-biotin-3′) was used as a hybridization probe. The sequence of the probe is complementary to A36–A14 of T. thermophilus tRNAGGUThr. The eluted tRNAs were purified further by 10% PAGE (7 m urea).
Nucleoside Analysis
Modified nucleosides were analyzed by HPLC (Hitachi L-2000 system) equipped with a reverse-phase C18 column (NUCLEOSIL 100 C18; 25 cm × 4.6 mm, 7 μm; GL Sciences, Inc.) as described previously (35).
RESULTS
Purification of T. thermophilus TrmI
The expression and purification of His-tagged TrmI from T. thermophilus HB27 were reported previously by Droogmans et al. (20). The amino acid sequences of the TrmI proteins from T. thermophilus strains HB8 and HB27 are identical. In the current study, we expressed TrmI protein from T. thermophilus HB8 without a His tag. Given that the solubility of TrmI protein was increased in the presence of KCl as reported (20, 23), we devised purification procedures that used relatively high concentrations of KCl (greater than 100 mm). We found that the combined use of heparin and gel filtration column chromatography was effective in maintaining high KCl concentrations during the purification as described under “Experimental Procedures.” As shown in Fig. 1, we successfully purified TrmI protein without a His tag.
FIGURE 1.
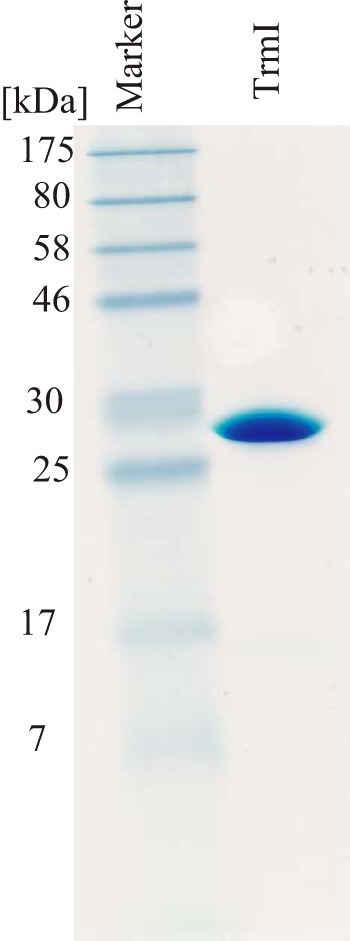
15% SDS-PAGE analysis of purified TrmI protein. An aliquot of 8 μg of purified TrmI was analyzed by 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue.
T. thermophilus TrmI Prefers to Methylate T. thermophilus tRNAPhe as Compared with S. cerevisiae tRNAPhe
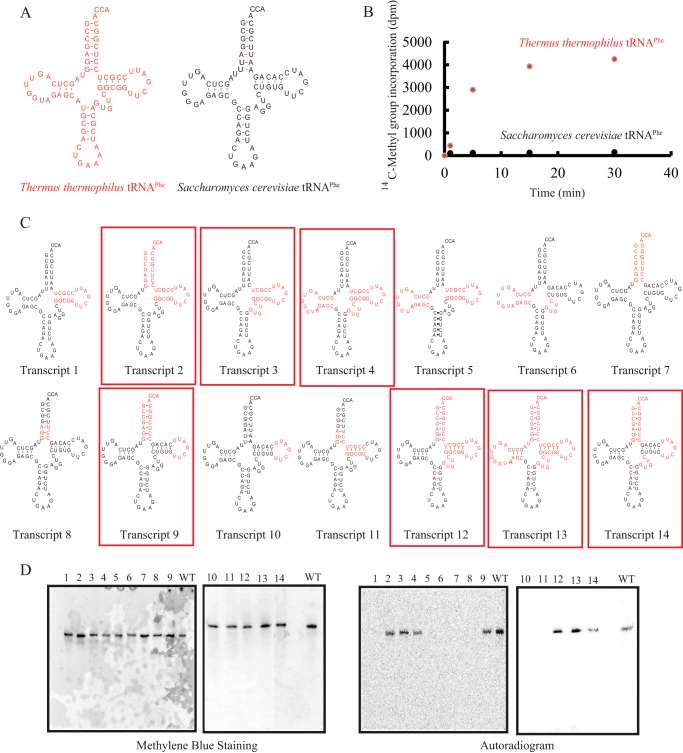
The tRNAPhe transcript from S. cerevisiae has been used as a model substrate for tRNA modification enzymes (36, 42–44) because its structure is well established (45–47). However, the purified TrmI from T. thermophilus methylated S. cerevisiae tRNAPhe very slowly (Fig. 2 and Table 1). When the methyl group acceptance activities of the tRNAPhe transcripts from T. thermophilus and S. cerevisiae (Fig. 2A) were compared, the initial velocity for the methyl group acceptance of S. cerevisiae tRNAPhe transcript was below 5% of that for the T. thermophilus tRNAPhe transcript (Fig. 2B). The kinetic parameters Km and Vmax for the T. thermophilus tRNAPhe transcript were determined to be 130 nm and 360 mmol mg−1 h−1, respectively (Table 1). These values are comparable with those obtained for other tRNA methyltransferases with tRNA transcripts (36, 44). Thus, the methylation of T. thermophilus tRNAPhe transcript by TrmI is not a special case. In contrast, we could not determine the kinetic parameters for the S. cerevisiae tRNAPhe transcript due to its low methyl group acceptance activity (Table 1). To clarify which features of the tRNA are responsible for this difference, we prepared 14 chimeric tRNA transcripts (Fig. 2C) and measured their methyl group acceptance activities (Fig. 2D and Table 1). In Fig. 2, the transcript numbers correspond to the lane numbers. When the T-arm of the S. cerevisiae tRNAPhe transcript was replaced with that of the T. thermophilus tRNAPhe transcript, the methyl group acceptance activity did not increase (Fig. 2D, lane 1). In contrast, when both the T-arm and aminoacyl stem of the S. cerevisiae tRNAPhe transcript were replaced with those of the T. thermophilus tRNAPhe transcript, the methyl acceptance activity increased clearly (Fig. 2D, lane 2). However, replacement of the aminoacyl stem alone did not have any effect on the methyl group acceptance activity (Fig. 2D, lane 7). These results show that the T-arm and aminoacyl stem in combination affect the methyl group acceptance activity. The replacement of both the aminoacyl stem and the T-loop in the S. cerevisiae tRNAPhe transcript also increased the methyl group acceptance activity (Fig. 2D, lane 9), which demonstrates that the T-loop and aminoacyl stem in combination are important for substrate tRNA recognition by TrmI. Similarly, the T-loop and variable region in combination were also important for substrate tRNA recognition by TrmI (Fig. 2D, lanes 3, 4, 12, 13, and 14). These results suggest that the sites involved in recognition by TrmI are the aminoacyl stem, T-loop, and variable region: their combined features are important for the efficient transfer of methyl groups.
FIGURE 2.
Chimeric tRNA transcripts of T. thermophilus and S. cerevisiae tRNAPhe. A, T. thermophilus tRNAPhe (left) and S. cerevisiae (right) tRNAPhe are depicted as clover leaf structures. To distinguish the two tRNAs, T. thermophilus tRNAPhe is indicated in red. B, the methyl group acceptance activities of T. thermophilus and S. cerevisiae tRNAPhe were compared. S. cerevisiae tRNAPhe was methylated very slowly by TrmI: the initial velocity of methyl group acceptance of S. cerevisiae tRNAPhe was ∼4% of that of T. thermophilus tRNAPhe. C, 14 chimeric tRNA transcripts are depicted as clover leaf structures. The sequences derived from T. thermophilus tRNAPhe are colored in red. The chimeric transcripts whose methyl group acceptance activities were more than 50% of that of T. thermophilus tRNAPhe are enclosed with red boxes. D, after methylation by TrmI and [methyl-14C]AdoMet, the chimeric tRNA transcripts (0.02 A260 unit each) were analyzed by 10% PAGE (7 m urea) and then visualized by methylene blue staining (left panels). The lane numbers correspond to the transcript numbers shown in C. WT indicates the T. thermophilus wild-type tRNAPhe transcript. To analyze the methyl group acceptance activities of the tRNA transcripts, the same gels were subjected to autoradiography (right panels). The kinetic parameters are summarized in Table 1.
TABLE 1.
Kinetic parameters for chimera tRNA transcripts
The relative Vmax/Km for the T. thermophilus wild-type tRNAPhe transcript was expressed as 100%. ND means that the kinetic parameters could not be calculated correctly.
| Transcript name | Km | Vmax | Relative Vmax/Km or relative initial velocity |
|---|---|---|---|
| nm | μmol mg−1 h−1 | % | |
| T. thermophilus | |||
| Wild-type tRNAPhe | 130 ± 20 | 360 ± 60 | 100 |
| S. cerevisiae | |||
| Wild-type tRNAPhe | ND | ND | <5a |
| Transcript 1 | ND | ND | <5a |
| Transcript 2 | 200 ± 40 | 450 ± 60 | 80 |
| Transcript 3 | 150 ± 40 | 360 ± 60 | 70 |
| Transcript 4 | 100 ± 20 | 260 ± 50 | 95 |
| Transcript 5 | >500 | <130 | 10 |
| Transcript 6 | ND | ND | <5a |
| Transcript 7 | ND | ND | <5a |
| Transcript 8 | ND | ND | <5a |
| Transcript 9 | 80 ± 20 | 200 ± 50 | 90 |
| Transcript 10 | ND | ND | <5a |
| Transcript 11 | ND | ND | <5a |
| Transcript 12 | 240 ± 50 | 710 ± 120 | 130 |
| Transcript 13 | 180 ± 50 | 660 ± 100 | 135 |
| Transcript 14 | 320 ± 60 | 450 ± 80 | 50 |
a The relative initial velocities were calculated from the time course experiment for 30 min. <5 means that the relative initial velocity was below 5% of that for T. thermophilus tRNAPhe.
Methyl Group Acceptance Activities of tRNA Deletion Variants
In general, tRNA methyltransferases recognize the local structure around the target site in the tRNA, including tertiary structural elements such as a stem-loop structure(s) (36, 48–51). Consequently, we prepared 10 deletion mutant transcripts of tRNAPhe from T. thermophilus (Fig. 3). The kinetic parameters for these transcripts are summarized in Table 2. In the previous study (49), we used two microhelices (Transcripts 15 and 16). Transcript 15 mimics the T-arm structure of E. coli tRNAPhe and was previously used for the crystallization of E. coli TrmA and RNA complex (52). Transcript 16 has an artificial sequence to reinforce the stem structure. In the previous study (49), it was confirmed that both Transcripts 15 and 16 were well methylated by T. thermophilus TrmFO (tRNA (m5U54) methyltransferase). In the current study, we tested whether Transcripts 15 and 16 were methylated by TrmI. 5.1 μm transcript, 0.1 μm TrmI, and 37 μm [methyl-14C]AdoMet were incubated at 55 °C for 30 min. However, Transcripts 15 and 16 were not methylated at all (Fig. 3B): the autoradiogram shown in Fig. 3B was obtained after a long exposure (3 days). We considered that some sequences in the T-arm might affect the methyl group acceptance activity. Therefore, next we tested the methyl group acceptance activity of Transcript 17, which mimics the T-arm of T. thermophilus tRNAPhe. Transcript 17 was methylated by TrmI very slowly (Fig. 3C and Table 2). The initial velocity of methyl transfer to Transcript 17 was below 5% of that to the full-length tRNA transcript. It should be mentioned that the results in Fig. 3C do not represent the relative initial velocities: the reaction mixture was incubated for 30 min, and the full-length (WT) tRNA transcript was near fully modified under this condition as shown in Fig. 2B. The difference between Transcripts 15 and 17 is only the T-stem sequence. The m1A58 modification is commonly found in T. thermophilus tRNAs (32–35, 41), and these tRNAs have different T-stem sequences. Consequently, these results suggested that TrmI recognizes the ribose phosphate backbone of T-stem. When the single-stranded extension corresponding to the 3′-side of acceptor stem was added to the T-arm (Transcript 18), the methyl group acceptance activity was significantly increased (Fig. 3D and Table 2). The data in Fig. 3, D and E, were obtained by incubations for 5 min. Consequently, the results approximated the relative initial velocities of methyl group acceptance of the transcripts. As shown in Fig. 3D and Table 2, Transcript 18 was efficiently methylated. Thus, the presence of the single-stranded extension corresponding to the 3′-side of acceptor stem is important for substrate RNA recognition by TrmI. When the variable region was added to Transcript 18 (Transcript 19), the methyl group acceptance activity was further increased (Fig. 3D and Table 2). Indeed, the initial velocity of methyl group acceptance of Transcript 19 was comparable with that of the full-length tRNA transcript (Fig. 3E and Table 2). Similarly, the transcripts that contained the T-arm structure with the aminoacyl stem or variable region were methylated by TrmI (Transcripts 20–24 in Fig. 3D and Table 2). However, Transcript 23, which did not have the aminoacyl stem, was methylated slowly. This result shows the importance of aminoacyl stem structure. Transcript 24 was methylated more rapidly than the wild-type tRNA transcript. This result suggests that the presence of the anticodon arm might disturb the methylation by TrmI. We determined the kinetic parameters for these transcripts (Table 2). The kinetic study showed the tendency that the deletion mutant transcripts have relatively large Km and Vmax values as compared with the wild-type tRNA transcript. This might be caused by the differences in the initial binding and structural change processes in the methyl transfer reaction. These results show that the combination of T-arm, variable region, and single-stranded extension correspond to the 3′-side of acceptor stem, which is consistent with the results from the chimeric transcripts shown in Fig. 2.
FIGURE 3.
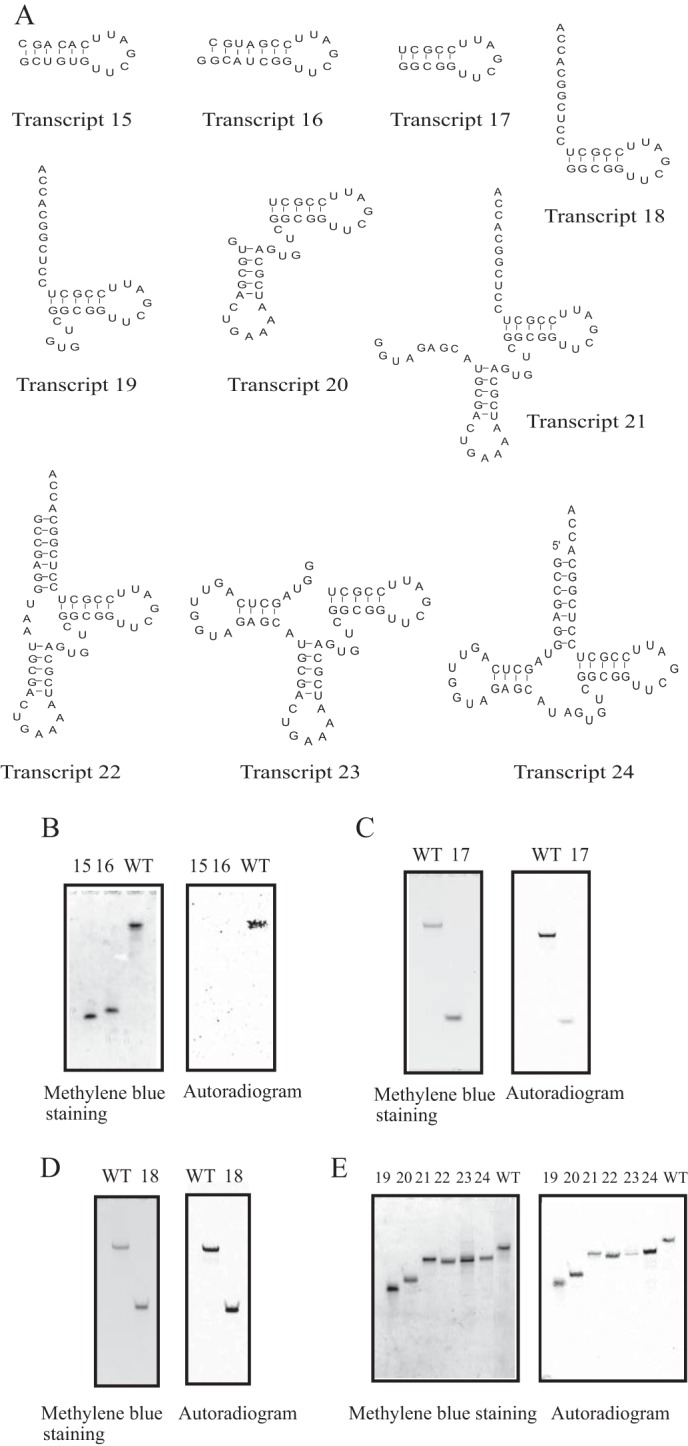
Deletion variants of T. thermophilus tRNAPhe. A, sequences of 10 deletion variants are shown. B, the methyl group acceptance activities of Transcripts 15 and 16 were checked. WT indicates the full-length T. thermophilus tRNAPhe transcript. C, the methyl group acceptance activity of Transcript 17 was tested. Very slow methyl transfer was observed in a long incubation for 30 min (right panel). D and E, the methyl group acceptance activities of Transcripts 18, 19, 20, 21, 22, 23, and 24 were checked with the gel assay. As shown in the autoradiogram (right panel), all tested transcripts were methylated by TrmI. The kinetic parameters are summarized in Table 2.
TABLE 2.
Kinetic parameters for deletion mutant transcripts
The relative Vmax/Km for the T. thermophilus wild-type tRNAPhe transcript was expressed as 100%. ND means that the kinetic parameters could not be calculated correctly. Not detectable means that the methyl group incorporation was not detectable under the tested condition.
| Transcript name | Km | Vmax | Relative Vmax/Km or relative initial velocity |
|---|---|---|---|
| nm | μmol mg−1 h−1 | % | |
| Full-length tRNAPhe | 130 ± 20 | 360 ± 60 | 100 |
| Transcript 15 | ND | ND | Not detectable |
| Transcript 16 | ND | ND | Not detectable |
| Transcript 17 | ND | ND | <5a |
| Transcript 18 | 280 ± 40 | 560 ± 90 | 75 |
| Transcript 19 | 200 ± 40 | 520 ± 70 | 95 |
| Transcript 20 | 200 ± 40 | 730 ± 120 | 135 |
| Transcript 21 | 220 ± 50 | 500 ± 60 | 85 |
| Transcript 22 | 250 ± 50 | 810 ± 100 | 120 |
| Transcript 23 | 270 ± 40 | 400 ± 90 | 25 |
| Transcript 24 | 130 ± 30 | 680 ± 70 | 190 |
a The relative initial velocities were calculated from the methyl group acceptance activities at 30-min periods. <5 means that the relative initial velocity was below 5% of that for T. thermophilus tRNAPhe.
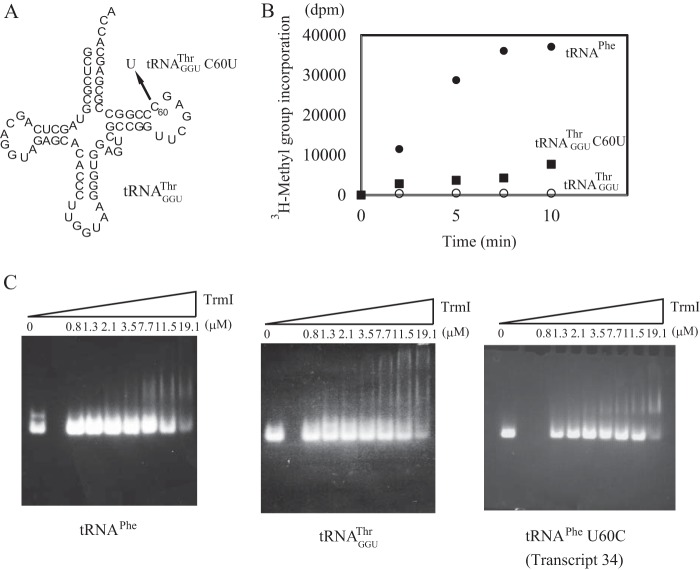
Disruption of T-stem Structure and G18-U55 and G19-C56 Tertiary Base Pairs
To determine whether the T-stem structure itself is required or not, we prepared a mutant transcript in which the T-stem was disrupted (Transcript 25 in Fig. 4A). This transcript was not methylated at all (Fig. 4B), which demonstrated that the stem structure is essential for recognition of the substrate by TrmI. The methyl group acceptance activities of the deletion mutants indicated that the tertiary interaction between the D- and T-arms was not required for recognition by TrmI (Fig. 3). To confirm this, we prepared a mutant transcript in which the G18G19 sequence was replaced with U18U19 (Transcript 26 in Fig. 4A). Interestingly, Transcript 26 was methylated more rapidly than the wild-type transcript (Fig. 4C). The kinetic parameters Km and Vmax for Transcript 26 were determined to be 150 nm and 480 μmol mg−1 h−1, respectively. Thus, the difference in methyl group acceptance activities between Transcript 26 and the wild-type transcript was mainly due to a difference in Vmax values. Given that the target site A58 forms a reverse Hoogsteen base pair with U54, A58 is embedded in the L-shaped tRNA structure. Consequently, the access of TrmI to A58 requires disruption of the tRNA structure. Disruption of the interaction between the T- and D-arms in Transcript 26 seemed to accelerate this structural change.
FIGURE 4.
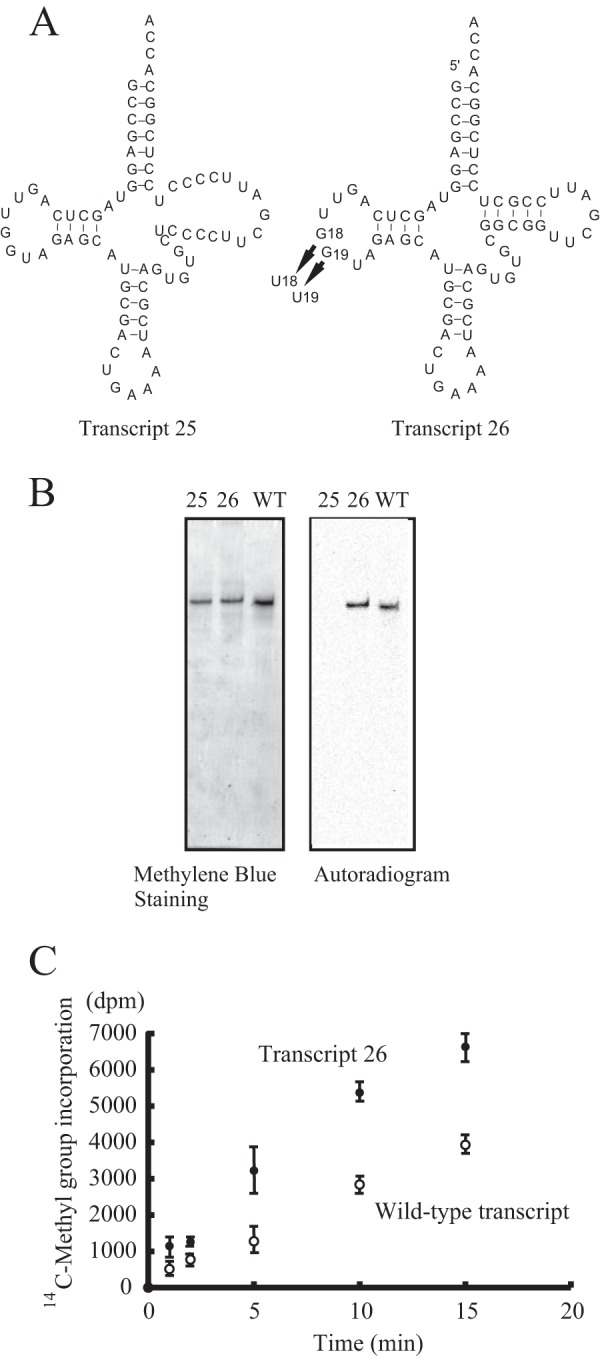
Disruptant tRNA mutants of T-stem and interaction between D- and T-arms. A, the T-stem of T. thermophilus tRNAPhe was disrupted by replacing the T-stem sequence (Transcript 25). The interaction between D- and T-arms was disrupted by replacing the G18G19 sequence with U18U19 (Transcript 26). B, the methyl group acceptance activities of the two disruptant tRNA mutants were analyzed with the gel assay. WT indicates the T. thermophilus wild-type tRNAPhe transcript. The lane numbers correspond to the transcript numbers. C, the methyl group acceptance activities of Transcript 26 and the wild-type transcript were compared. Error bars represent S.E.
Introduction of Point Mutations Revealed That U55, C56, U59, and U60 Are Involved in Recognition of the Substrate tRNA by TrmI
To clarify the positive sequence determinants in the tRNA for methylation by TrmI, we prepared an additional 17 tRNAPhe mutant transcripts with a single point mutation introduced in each (Transcripts 27–43 in Fig. 5). The kinetic parameters are given in Table 3. The point mutations were introduced mainly into the T-loop. Given that several tRNAs (for example tRNAfMet) have an A at position 46 instead of G (32), we prepared one mutant tRNA transcript in which G46 was replaced by A (Transcript 27). This transcript was methylated efficiently by TrmI (Fig. 5B). When U54 was replaced with A (Transcript 28), G (Transcript 35), or C (Transcript 36), the methyl group acceptance activity was not changed significantly, suggesting that the U54-A58 reverse Hoogsteen base pair is not required for the methylation by TrmI. When U55 was replaced with A (Transcript 29), the methyl group acceptance activity was decreased considerably, whereas if it was replaced with G (Transcript 37) or C (Transcript 38), the activity was not changed significantly. These results might be explained by the formation of a Watson-Crick base pair between A55 and U59 in the Transcript 29. When C56 was replaced with G (Transcript 30), U (Transcript 39), or A (Transcript 40), the methyl group acceptance activity was lost completely, which showed that C56 is a positive determinant for methylation by TrmI. The replacement of G57 with C (Transcript 31) decreased the methyl group acceptance activity considerably, whereas its replacement with A (Transcript 41) had no effect on the activity. These results show that a purine at position 57 is a positive determinant for methylation by TrmI. The replacement of U59 with C (Transcript 33) decreased the methyl group acceptance activity considerably, whereas its replacement with G (Transcript 42) had no significant effect on the activity. Although the precise mechanism of this phenomenon cannot be explained, the replacement of U59 with C might change the structure of ribose phosphate backbone of the T-loop. Furthermore, when U60 was replaced with C (Transcript 34) or G (Transcript 43), the methyl group acceptance activity was significantly decreased. These results are consistent with the low methyl group acceptance activity of the S. cerevisiae tRNAPhe transcript, which has a C at position 60 (Fig. 2A). Thus, U60 is also a positive determinant for methylation by TrmI. From these results, the preferred T-loop sequence for methylation by T. thermophilus TrmI can be derived (Fig. 5C).
FIGURE 5.
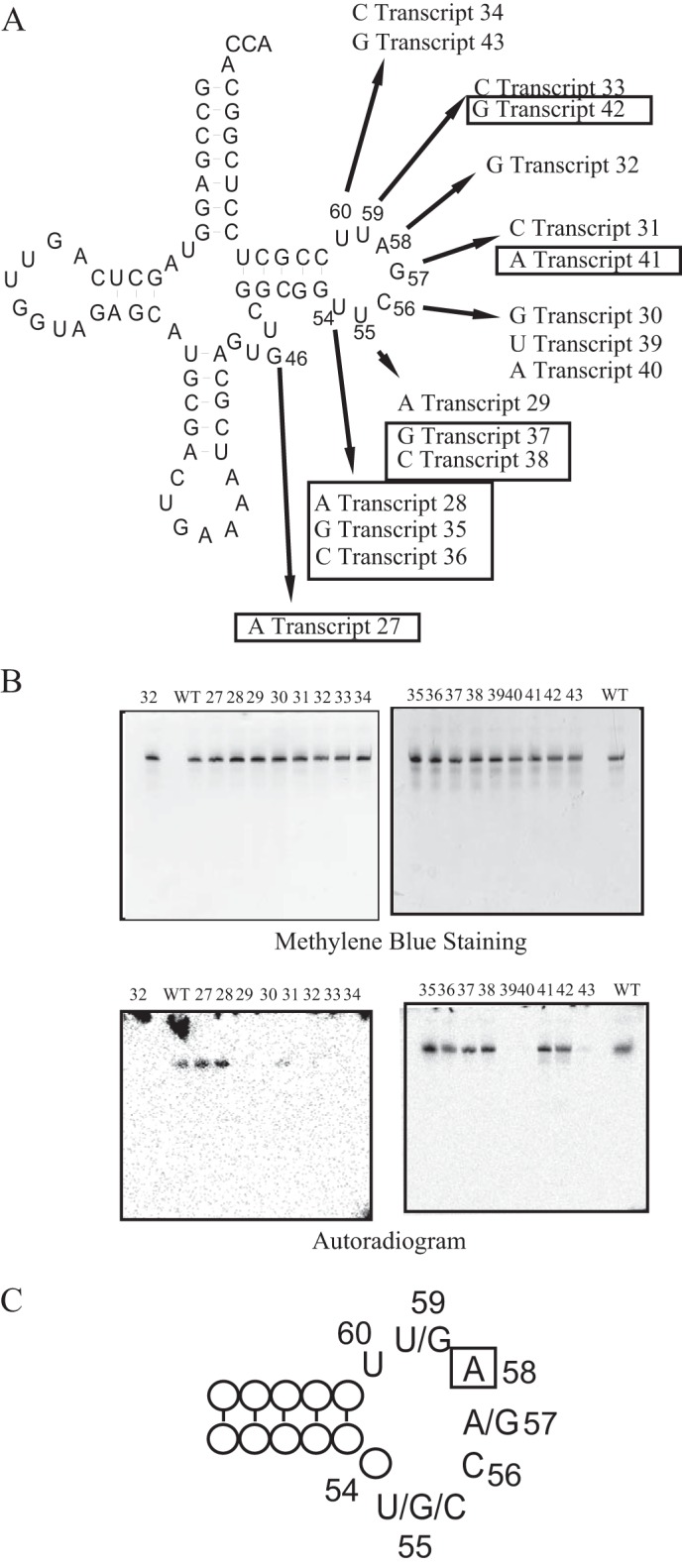
Point mutations in the T. thermophilus tRNAPhe transcript. A, 17 point mutations were introduced individually into the T. thermophilus tRNAPhe transcript. The arrows show the mutations and transcript numbers. The mutation sites and positions are indicated in the clover leaf structure. The transcripts of which the initial velocities of methyl group acceptance activity were more than 50% of that of the wild-type tRNAPhe transcript are enclosed with boxes. The kinetic parameters are summarized in Table 3. B, the methyl group acceptance activities of point mutants were analyzed with the gel assay. The upper panels show the gel stained with methylene blue. The lane numbers correspond to the transcript numbers. WT indicates the T. thermophilus wild-type tRNAPhe transcript. Incorporation of the [14C]methyl group was monitored by autoradiography of the same gels (lower panels). C, the preferred T-loop sequence for methylation by TrmI is depicted. The methylation site A58 is enclosed with a box.
TABLE 3.
Kinetic parameters for point mutated tRNA transcripts
The relative Vmax/Km for the T. thermophilus wild-type tRNAPhe transcript was expressed as 100%. ND means that the kinetic parameters could not be calculated correctly.
| Transcript name | Mutation site | Km | Vmax | Relative Vmax/Km or relative initial velocity |
|---|---|---|---|---|
| nm | μmol mg−1 h−1 | % | ||
| T. thermophilus wild-type tRNAPhe | None | 130 ± 20 | 360 ± 60 | 100 |
| Transcript 27 | 46G→A | 110 ± 20 | 340 ± 60 | 110 |
| Transcript 28 | 54U→A | 130 ± 30 | 440 ± 70 | 125 |
| Transcript 29 | 55U→A | ND | ND | <5a |
| Transcript 30 | 56C→G | ND | ND | Not detectable |
| Transcript 31 | 57G→C | ND | ND | <10a |
| Transcript 32 | 58A→G | ND | ND | Not detectable |
| Transcript 33 | 59U→C | ND | ND | <5a |
| Transcript 34 | 60U→C | ND | ND | <5a |
| Transcript 35 | 54U→G | 120 ± 20 | 480 ± 80 | 150 |
| Transcript 36 | 54U→C | 130 ± 40 | 250 ± 60 | 80 |
| Transcript 37 | 55U→G | 100 ± 20 | 380 ± 80 | 135 |
| Transcript 38 | 55U→C | 120 ± 20 | 390 ± 70 | 120 |
| Transcript 39 | 56C→U | ND | ND | Not detectable |
| Transcript 40 | 56C→A | ND | ND | Not detectable |
| Transcript 41 | 57G→A | 130 ± 20 | 480 ± 60 | 150 |
| Transcript 42 | 59U→G | 160 ± 30 | 340 ± 60 | 80 |
| Transcript 43 | 60U→G | ND | ND | <5a |
a The relative initial velocities were calculated from the time course experiment for 30 min. <5 and <10 mean that the relative initial velocity was below 5% and 10% of that for T. thermophilus tRNAPhe, respectively.
Replacement of A58 by I Causes the Complete Loss of Methyl Group Acceptance Activity
The replacement of A58 by G caused the complete loss of methyl group acceptance activity (Transcript 32). This result shows that TrmI correctly recognizes an adenine base at position 58. To clarify which features of the structure of adenine are recognized by TrmI, we prepared a mutant tRNAPhe transcript in which A58 was replaced with I (Transcript 44) or 2-AP (Transcript 45). Hypoxanthine (the base in inosine) and 2-AP have a structure very similar to that of adenine except that it does not contain an amino group at position 6 (Fig. 6A). As described under “Experimental Procedures,” the 3′-half of tRNA containing I or 2-AP at position 58 was synthesized chemically and ligated with the 5′-half using T4 RNA ligase (Fig. 6B). The ligated sample was purified further by 10% PAGE (7 m urea). As shown in Fig. 6C, these transcripts were not methylated at all, demonstrating that TrmI recognizes the 6-amino group in A58. During the revision of this manuscript, Hamdane et al. (22) reported that 2-AP in minihelices, which mimic the T-arm and aminoacyl-stem, inhibit the methylation by aTrmI. Thus, our experimental results are in good agreement with their results.
FIGURE 6.
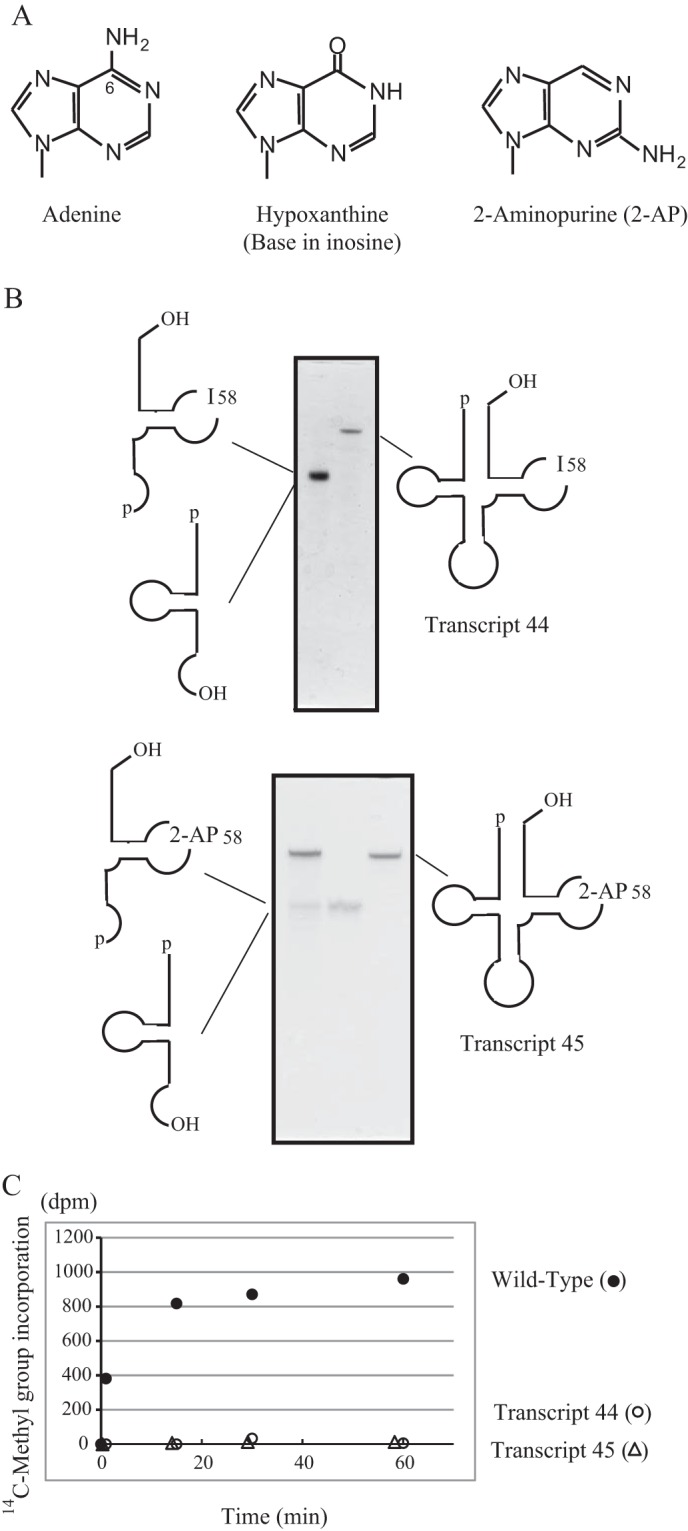
TrmI recognizes the 6-amino group in A58. A, the structures of adenine, hypoxanthine, and 2-aminopurine bases are compared. Position 6 of adenine is numbered. B, T. thermophilus tRNAPhe mutants that contained I58 and 2-AP were constructed by RNA ligation. The resultant ligated samples (Transcripts 44 and 45) were purified by 10% PAGE (7 m urea). The gel was stained with methylene blue. The bands that corresponded to 5′- and 3′-halves of tRNAPhe overlapped on the gel. The left lane of the lower panel shows the ligation sample before the purification. C, the [14C]methyl group acceptance activities of Transcripts 44 (open circles) and 45 (open triangles) were tested. Filled circles show the methyl group acceptance activity of the wild-type tRNAPhe.
Transfer RNAGGUThr from T. thermophilus Has a C at Position 60
In general, the base at position 60 in tRNA is conserved as a pyrimidine (10). However, TrmI prefers U60 to C60 as shown in Fig. 5C. T. thermophilus has 47 tRNA species. Among them, only tRNAGGUThr contains C60 instead of U60 (Fig. 7A). Given that the anticodons of the other two tRNAThr species are CGU and UGU, tRNAGGUThr is required to decode ACC and ACU threonine codons during protein synthesis. To check the methyl group acceptance activity of tRNAGGUThr, we prepared the tRNAGGUThr transcript. As expected, the methyl group acceptance activity of the tRNAGGUThr transcript was considerably lower than that of the tRNAPhe transcript: the initial velocity of the former was only 2% of the latter (Fig. 7B). The replacement of C60 in the tRNAGGUThr transcript with U increased the initial velocity of methyl group acceptance activity by ∼6-fold (Fig. 7B). Thus, the presence of C60 is one of the reasons for the low methyl group acceptance activity of tRNAGGUThr. We thought that TrmI might have a lower affinity for tRNAGGUThr than for tRNAPhe. To confirm this, we performed a gel mobility shift analysis. As reported by Droogmans et al. (20), it is much more difficult to perform gel mobility shift assays with TrmI than with other tRNA modification enzymes (53, 54) because the large TrmI tetramer (more than 100 kDa) does not migrate into a normal polyacrylamide gel under electrophoresis. Consequently, we used agarose gels for the gel mobility shift assay as reported (20) and stained them with ethidium bromide. As shown in Fig. 7C, left panel, a discrete shifted band derived from the tRNAPhe transcript was clearly detected. In contrast, the band derived from the tRNAGGUThr transcript was smeared (Fig. 7C, middle panel). These results showed that the low methyl group acceptance activity of the tRNAGGUThr transcript is caused by a low affinity for TrmI. The presence of C60 did not affect the pattern obtained with the gel mobility shift assay significantly because the mutant tRNAPhe 60U→C (Fig. 7C, lower right panel) transcript gave similar shift patterns as the wild-type tRNA transcript. Thus, the high affinity of the tRNAPhe transcript seems to be caused by a combination(s) of aminoacyl stem structure, sequence of the T-loop, and variable region of tRNAPhe.
FIGURE 7.
The methyl group acceptance activity of tRNAGGUThr and its affinity for TrmI. A, among the 47 tRNA species in T. thermophilus, tRNAGGUThr is the only one that has a C at position 60. A mutant tRNA transcript was generated (tRNAGGUThr 60C→U) in which C60 was replaced with U. B, the methyl group acceptance activities of the wild-type tRNAGGUThr (open circles) and 60C→U mutant (filled squares) were compared with that of tRNAPhe (filled circles). The initial velocity of incorporation of [3H]methyl group into the wild-type tRNAGGUThr was 2% of that of tRNAPhe. The replacement of C60 with U accelerated the velocity of incorporation of [3H]methyl group. C, the affinities of transcripts for TrmI were analyzed by the gel mobility shift assay.
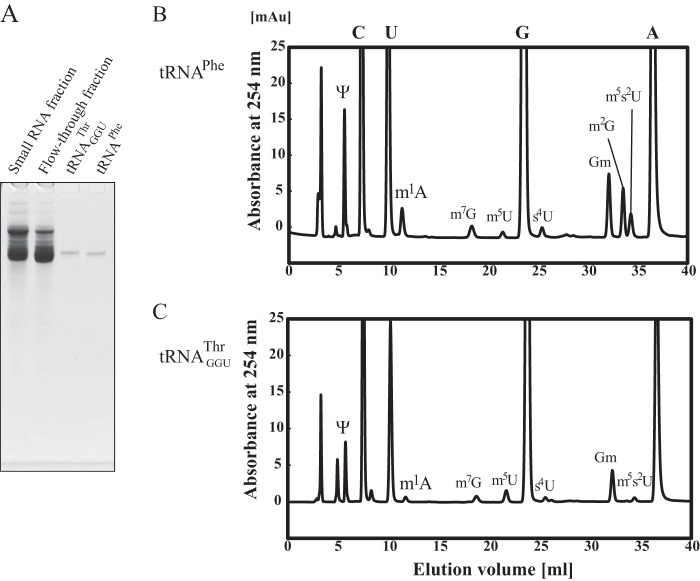
The Other Modifications in tRNAGGUThr Accelerate the Methylation by TrmI
Recently, we reported that the tRNA modification enzymes and modified nucleotides in T. thermophilus form a network, which regulates the extent of modifications in tRNA in response to changes in temperature (2, 34, 35, 51, 55). The m1A58 modification by TrmI is required for cell viability at high temperatures (20) because it accelerates the sulfur transfer reaction that is required for m5s2U54 modification (37). The m5s2U54 modification increases the melting temperature of the tRNA by more than 4 °C (32, 33). Furthermore, the combination of the Gm18, m5s2U54, and m1A58 modifications increases the melting temperature by nearly 10 °C as compared with the unmodified transcript (35). The m1A58 modification is a relatively late modification in T. thermophilus tRNA. Consequently, we proposed that the other modifications in tRNAGGUThr might support the formation of m1A58 by TrmI. To confirm this idea, we utilized a T. thermophilus HB8 trmI gene disruptant strain (37). Given that this HB8 gene disruptant strain did not grow at 80 °C (37) as reported with the analogous HB27 strain (20), the cells were cultured at 70 °C. We prepared the fraction of small RNAs (mainly tRNA and 5 S rRNA) from the wild-type and trmI gene disruptant strains and analyzed their modified nucleosides (Fig. 8A). The small RNA fraction from the wild-type strain contains the m1A (Fig. 8A, upper panel). In contrast, the small RNA fraction from the trmI gene disruptant strain did not contain the m1A nucleoside (Fig. 8A, middle panel). The elution point of m1A was confirmed with the standard marker (Fig. 8A, lower panel). As shown in Fig. 8B, tRNAGGUThr was purified successfully from the trmI gene disruptant strain by the solid-phase DNA probe method (40, 41). The methyl transfer assay with TrmI revealed that this tRNA was methylated more rapidly than the tRNAGGUThr transcript (Fig. 8C), which demonstrated that the other modifications present in tRNAGGUThr accelerated the methylation by TrmI. However, the initial velocity of methyl group acceptance activity of tRNAGGUThr from the trmI gene disruptant strain was only 13% of that of the tRNAPhe transcript (Fig. 8C).
FIGURE 8.
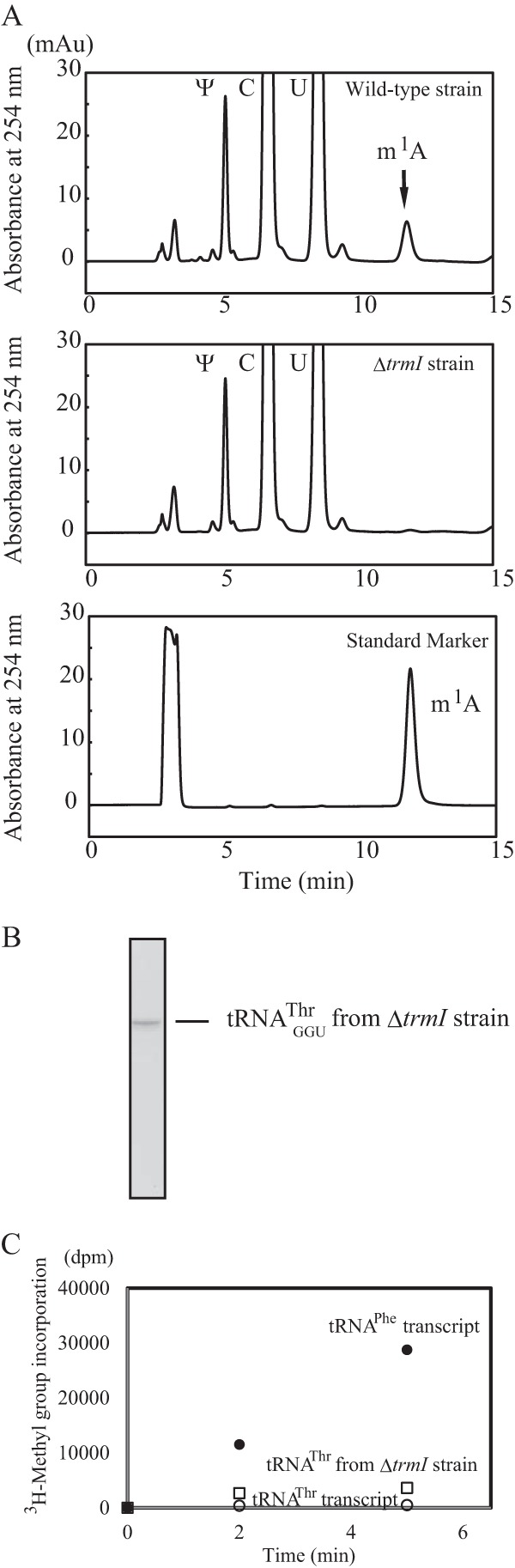
tRNAGGUThr purified from the trmI gene disruptant strain is methylated more rapidly by TrmI than the tRNAGGUThr transcript. A, the modified nucleosides in the fraction of small RNA from T. thermophilus wild-type (upper panel) and trmI gene disruptant (middle panel) strains were compared. The lower panel shows the elution point of the standard m1A marker. B, transfer RNAGGUThr was purified from the trmI gene disruptant strain by the solid-phase DNA probe method. 0.02 A260 unit of tRNAGGUThr was analyzed by 10% PAGE (7 m urea). The gel was stained with methylene blue. C, the methyl group acceptance activity of tRNAGGUThr from the trmI gene disruptant strain (squares) was compared with those of the tRNAPhe (filled circles) and tRNAGGUThr (open circles) transcripts. The presence of modified nucleotides in purified tRNAGGUThr accelerated the methylation by TrmI. mAu, milli-absorbance units.
In Living Cells, tRNAGGUThr Is Not Modified fully by TrmI
We thought that tRNAGGUThr might not be modified fully in living cells because the methyl group acceptance activity of tRNAGGUThr purified from the trmI gene disruptant strain was rather low as shown in Fig. 8C. Consequently, we investigated the m1A content in tRNAGGUThr from the wild-type strain. It should be mentioned that the content of m1A in tRNA changes depending on the culture temperature, components of the medium, and air supply (35, 55, 56). In this experiment, the cells were cultured at 70 °C in rich medium with vigorous shaking. Under these conditions, more than 85% of tRNAPhe contained the m1A58 modification (41). To estimate the m1A content in tRNAGGUThr, we purified tRNAPhe and tRNAGGUThr from the wild-type strain (Fig. 9A). The modified nucleosides in tRNAPhe (Fig. 9B) and tRNAGGUThr (Fig. 9C) were compared. The levels of m1A were calculated from the peak areas of m1A and pseudouridine (Ψ): tRNAPhe contains two Ψ residues (Ψ39 and Ψ55), whereas tRNAGGUThr contains one Ψ residue (Ψ55). When the level of Ψ in tRNAGGUThr was set as 1.00, the level of the m1A modification in this tRNA was calculated to be 0.39. In contrast, when the level of Ψ in tRNAPhe was set as 2.00, the level of m1A was calculated to be 0.90. These calculations were based on the assumption that the Ψ residues were fully modified in these tRNAs. Therefore, we tested the level of m1A58 modification by the other method: the purified tRNAPhe and tRNAGGUThr were fully methylated by TrmI and [methyl-14C]AdoMet. The levels of the m1A modification in tRNAPhe and tRNAGGUThr were calculated to be 93 and 48%, respectively. These results clearly showed that tRNAGGUThr is not modified fully in the living cells cultured at 70 °C even though the other modifications present in the tRNA accelerate the formation of m1A58 by TrmI. Furthermore, the level of m5s2U in tRNAGGUThr was low as compared with that in tRNAPhe, and the level of an intermediate, m5U, in tRNAGGUThr was increased (Fig. 9, B and C). This observation is consistent with the previous proposed network (2, 55). Moreover, the levels of m7G, s4U, and Gm in tRNAGGUThr were considerably low as compared with those in tRNAPhe (Fig. 9, B and C). These differences might be caused by the low levels of m1A and m5s2U in tRNAGGUThr.
FIGURE 9.
Nucleoside analysis of tRNAPhe and tRNAGGUThr from T. thermophilus wild-type strain. To estimate the m1A content in the tRNAGGUThr fraction, tRNAPhe and tRNAGGUThr were purified from the T. thermophilus wild-type strain, and their modified nucleosides were compared. A, the purified tRNAs were analyzed by 10% PAGE (7 m urea). The gel was stained with toluidine blue. The modified nucleosides in tRNAPhe (B) and tRNAGGUThr (C) were analyzed. mAu, milli-absorbance units.
DISCUSSION
Fig. 10 shows the crystal structures of TrmI and S. cerevisiae tRNAPhe. Given that an L-shaped tRNA cannot be simply placed to the enzyme surface of the crystal structure of TrmI without a clash between the two structures (23, 27), there is no proposed docking model between TrmI and tRNA. Thus, the structure of tRNA is expected to be disrupted during methylation by TrmI. Our experiments with the chimeric transcripts and deletion mutants showed that combinations of the aminoacyl stem, variable region, and T-loop of tRNA are required for efficient methyl transfer by TrmI: these regions are colored in gray (Fig. 10). The earlier crystal structure study reported the presence of two grooves (grooves 1 and 2), which include many basic amino acid residues (Fig. 10, blue regions), on the surface of the TrmI tetramer (23). The size and orientation between grooves and the catalytic pocket are consistent with a model in which the grooves capture the aminoacyl stem in the L-shaped tRNA (23). Indeed, our current study demonstrates that the aminoacyl stem is very important for the tRNA recognition by TrmI. The stem structure itself of the aminoacyl stem is not required because Transcript 18 in Fig. 3 was methylated efficiently. When the aminoacyl stem is placed on a groove, the anticodon arm cannot be placed on another groove without a structural clash (23). The results of our experiments with the chimeric transcripts and deletion mutants are consistent with their observation because the anticodon arm was not recognized by TrmI. Instead of the anticodon arm, another groove might capture the variable region because the combination of the aminoacyl stem, variable region, and T-arm was required for efficient methyl transfer by TrmI. Under physiological conditions, one TrmI tetramer seems to capture one tRNA because grooves 1 and 2 are formed by the tetrameric structure (23). This idea was reinforced by the gel shift pattern shown in Fig. 7: the tRNAPhe transcript, at least, yielded only a single shifted band. Given that the modification site A58 is located in the T-loop, the aminoacyl stem and/or variable region seems to be required for the initial binding process. The T-stem structure itself was required for methylation by TrmI. This result shows that the structure of the ribose phosphate backbone of the T-stem is required, which suggests that the distance and angle between the target site A58 and the initial binding sites (aminoacyl stem and variable region) are important for the methylation by TrmI. Given that the gel mobility shift assays with the tRNAPhe variant showed that the presence of a C or U at position 60 had no obvious effect on the formation of a complex between tRNA and TrmI, the C nucleotide at position 60 might be involved in the process of structural change. The precise order in which C60 and C56 (important positive sequence determinants) are recognized is unclear. However, it is clear that the recognition of C56 occurred after the disruption of T- and D-arm interaction because C56 forms a tertiary base pair with G19. Disruption of the interaction between the T- and D-arms accelerated the methyl transfer reaction by TrmI, suggesting that the methyl transfer reaction requires the disruption of the T- and D-arm interaction. Given that the target site A58 forms a reverse Hoogsteen base pair with U54 and is embedded in the tRNA structure, introduction of A58 into the catalytic pocket requires at least the disruption of the structure of the T-arm. Disruption of the T- and D-arm interaction is probably part of this process of structural change. The U54-A58 reverse Hoogsteen base pair is not required for the recognition of A58 because point mutation of U54 did not change the methyl group acceptance activity significantly. However, the 6-amino group in A58 is recognized by TrmI because the mutant tRNA transcripts that contained I58 and 2-AP58 were not methylated at all. The earlier crystal structure study proposed a hypothetical interaction between the 6-amino group in A58 and the catalytic center (aspartate 170 in T. thermophilus TrmI) (23). Our result in the current study reinforces their hypothetical catalytic mechanism: the 6-amino group in A58 is absolutely required for the methyl transfer reaction. In the archaeal modification system, aTrmI methylates both A57 and A58 (21), and the resultant m1A57 is further modified to m1I57 by deamination (57). Unmethylated I57 has not been found thus far in archaeal tRNAs. Furthermore, the crystal structure of aTrmI (26) strongly suggests that TrmI and aTrmI have the same catalytic mechanism. These observations suggest that aTrmI also recognizes the 6-amino group in the target adenosine similarly to eubacterial TrmI. Indeed, during the revision of this manuscript, Hamdane et al. (22) reported that 2-AP inhibited the methylation of minihelices, which mimic the T-arm and aminoacyl stem, by aTrmI. Furthermore, they proposed the hypothetical mechanism that aTrmI methylates two adenosines, A57 and A58, without release of the intermediate (m1A57-modified tRNA) (22). Thus, the reaction mechanism of aTrmI seems to be different from that of Trm1, although these enzymes consume two AdoMet molecules in the reaction: Trm1 releases the intermediate (m2G26-modified tRNA) in the reaction (58, 59).
FIGURE 10.
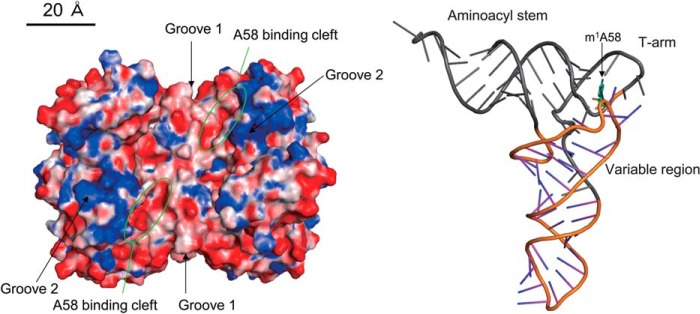
Structures of T. thermophilus TrmI and tRNA. The crystal structures of TrmI and S. cerevisiae tRNAPhe are depicted in the same scale. The acidic and basic residues in TrmI are colored in red and blue, respectively. The A58 binding clefts are enclosed by green circles. The aminoacyl stem, T-arm, and variable region in tRNA are colored in gray: these regions are important for the recognition by TrmI. The m1A58 is highlighted by a stick model (green).
For a long time, it has been thought that eubacterial TrmI modifies all tRNAs at the same efficiency. The results of our current study contradict this assumption. TrmI from T. thermophilus at least methylated tRNAPhe transcript 50 times faster than the tRNAGGUThr transcript. Indeed, more than half of native tRNAGGUThr in cells was not methylated, whereas 90% of tRNAPhe was modified. The network among modified nucleosides and modification enzymes has mainly been investigated by examining the modification patterns of mixtures of tRNA and of tRNAPhe (35, 55). However, the acceleration of formation of m5s2U54 in the presence of m1A58 is applicable even in the case of tRNAGGUThr (Fig. 9). The nucleotide at position 60 of tRNA is semiconserved as a pyrimidine. Among the 47 tRNA species from T. thermophilus, only tRNAGGUThr has a C at position 60 rather than a U. The existence of this tRNA would appear to be disadvantageous for growth at high temperatures because the m1A58 modification is required for introduction of the m5s2U54 modification at high temperatures (20, 37). Why does T. thermophilus have this apparently disadvantageous tRNA for survival at high temperatures? There are several possibilities. The first possibility is that the precursor tRNAs have different sequences, and their structural flexibilities are also different. Therefore, each tRNA transcript is modified in the different levels for the regulation of appropriate flexibilities at suitable temperatures (for example 70 °C). The second possibility is that T. thermophilus lives in hot springs, and organic compounds such as amino acids in hot springs are limited. Therefore, rapid speed in protein synthesis might be not required for survival under the poor nutrient environment. Because 40–50% of tRNAGGUThr contains the m1A58 modification even at 80 °C, the protein synthesis is not paused completely. The third possibility is that at high temperatures T. thermophilus produces heat-shock proteins (55). The population change of tRNA species might regulate the production of heat-shock proteins. The fourth possibility is that the C60 in tRNAGGUThr was derived from the accidental mutation, and it does not have any positive function for survival. Although there are these possibilities, the biological function of C60 in tRNAGGUThr is unclear. Recently, we measured the quantity of tRNAGGUThr in the small RNA fraction (41): the amount of tRNAGGUThr was only 0.25–0.30 A260 unit in the 50.0 A260 units of small RNA fraction. Thus, tRNAGGUThr seems to be a minor tRNA in T. thermophilus.
The requirement for a U at position 60 for efficient methylation by T. thermophilus TrmI seems to be applicable to the other thermophilic eubacterial TrmI enzymes. For example, all 43 tRNA species encoded in the genome of Aquifex aeolicus, a hyperthermophilic eubacterium, have the U60 nucleotide (10). In contrast, tRNA species with a C at position 60 are often found in mesophilic eubacteria. For example, three tRNA species in E. coli and seven tRNA species in Bacillus subtilis contain C60 (10): these mesophilic eubacteria do not have the TrmI protein. Therefore, the presence of TrmI in thermophilic eubacteria might function as a selective pressure for the evolution of tRNA sequences. It should be mentioned that these discussions about TrmI are only applicable to eubacterial tRNAs because in eukaryotes the m1A modification is generated by a different enzyme (the Trm6-Trm61 complex). In fact, 20 of 51 tRNA species from S. cerevisiae have a C at position 60, and these tRNAs contain the m1A58 modification (10). Interestingly, the U60 nucleotide is semiconserved among archaeal tRNAs: there are very few tRNAs that have a C at this position. The semiconservation of U60 in archaeal tRNA is observed in both thermophiles and mesophiles (10). Therefore, the requirement for U60 for efficient methylation might also be applicable to aTrmI enzymes.
In thermophilic eubacteria, the m1A58 modification functions as a tRNA stabilization factor and part of the tRNA modification network. However, in the case of eukaryotes, the m1A58 modification functions as a part of the RNA quality control system (4). Precursor initiator tRNAMet that does not contain the m1A58 modification is polyadenylated by the so-called “TRAMP complex” and then degraded by Rrp6 and the nucleosome (4). Furthermore, human immunodeficiency virus utilizes the m1A58 modification in human tRNA3Lys as the terminator of reverse transcription (7). Therefore, studies on the formation of m1A58 in eukaryotic tRNA are very important for understanding cellular biological phenomena and the control of infectious organisms. However, the enzyme architectures of eukaryotic Trm6-Trm62 and eubacterial TrmI are different. Several infectious eubacteria such as Mycobacterium tuberculosis contain a TrmI protein (20, 24). Therefore, eubacterial TrmI might be an effective target for anti-infective bacterial drugs. Thus, the studies on the m1A58 modification in eubacterial tRNA are also important.
This work was supported in part by Grant-in-aid for Science Research 23350081 (to H. H.) from the Japan Society for the Promotion Science.
- m1A
- 1-methyladenosine
- m1A58
- N1-methyladenosine at position 58
- aTrmI
- archaeal TrmI
- AdoMet
- S-adenosyl-l-methionine
- 2-AP
- 2-aminopurine
- Ψ
- pseudouridine.
REFERENCES
- 1. Machnicka M. A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K. M., Helm M., Bujnicki J. M., Grosjean H. (2013) MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hori H. (2014) Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 5, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phizicky E. M., Hopper A. K. (2010) tRNA biology charges to the front. Genes Dev. 24, 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaneko T., Suzuki T., Kapushoc S. T., Rubio M. A., Ghazvini J., Watanabe K., Simpson L., Suzuki T. (2003) Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J. 22, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takano Y., Takayanagi N., Hori H., Ikeuchi Y., Suzuki T., Kimura A., Okuno T. (2006) A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol. Microbiol. 60, 81–92 [DOI] [PubMed] [Google Scholar]
- 7. Renda M. J., Rosenblatt J. D., Klimatcheva E., Demeter L. M., Bambara R. A., Planelles V. (2001) Mutation of the methylated tRNALys3 residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J. Virol. 75, 9671–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gehrig S., Eberle M.-E., Botschen F., Rimbach K., Eberle F., Eigenbrod T., Kaiser S., Holmes W. M., Erdmann V. A., Sprinzl M., Bec G., Keith G., Dalpke A. H., Helm M. (2012) Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jöckel S., Nees G., Sommer R., Zhao Y., Cherkasov D., Hori H., Ehm G., Schnare M., Nain M., Kaufmann A., Bauer S. (2012) The 2′-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jühling F., Mörl M., Hartmann R. K., Sprinzl M., Stadler P. F., Pütz J. (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong R. S., Scarborough G. A., Borek E. (1971) Transfer ribonucleic acid methylases during the germination of Neurospora crassa. J. Bacteriol. 108, 446–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McFarlane E. S. (1972) Bases methylated in vitro by cell-free extract of adenovirus-18-induced tumours. Biochem. J. 129, 513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klagsbrun M. (1973) Differences in the methylation of transfer ribonucleic acid in vitro by the mitochondrial and cytoplasmic transfer ribonucleic acid methylases of HeLa cells. J. Biol. Chem. 248, 2606–2611 [PubMed] [Google Scholar]
- 14. Glick J. M., Leboy P. S. (1977) Purification and properties of tRNA(adenine-1)-methyltransferase from rat liver. J. Biol. Chem. 252, 4790–4795 [PubMed] [Google Scholar]
- 15. Kraus J. (1978) Recognition of individual Escherichia coli transfer ribonucleic acids by 1-adenine-specific methyltransferase from rat liver. Biochem. J. 169, 247–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wierzbicka H., Jakubowski H., Pawelkiewicz J. (1975) Transfer RNA methyltransferases from yellow lupin seeds: purification and properties. Nucleic Acids Res. 2, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mutzel R., Malchow D., Meyer D., Kersten H. (1986) tRNA (adenine-N1)-methyltransferase from Dictyostelium discoideum. Purification, characterization and developmental changes in activity. Eur. J. Biochem. 160, 101–108 [DOI] [PubMed] [Google Scholar]
- 18. Anderson J., Phan L., Cuesta R., Carlson B.A., Pak M., Asano K., Björk G.R., Tamame M., Hinnebusch A.G. (1998) The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12, 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anderson J., Phan L., Hinnebusch A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 5173–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Droogmans L., Roovers M., Bujnicki J. M., Tricot C., Hartsch T., Stalon V., Grosjean H. (2003) Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res. 31, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roovers M., Wouters J., Bujnicki J. M., Tricot C., Stalon V., Grosjean H., Droogmans L. (2004) A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 32, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamdane D., Guelorget A., Guérineau V., Golinelli-Pimpaneau B. (2014) Dynamics of RNA modification by a multi-site-specific tRNA methyltransferase. Nucleic Acids Res. 42, 11697–11706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barraud P., Golinelli-Pimpaneau B., Atmanene C., Sanglier S., Van Dorsselaer A., Droogmans L., Dardel F., Tisné C. (2008) Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J. Mol. Biol. 377, 535–550 [DOI] [PubMed] [Google Scholar]
- 24. Gupta A., Kumar P. H., Dineshkumar T. K., Varshney U., Subramanya H. S. (2001) Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 312, 381–391 [DOI] [PubMed] [Google Scholar]
- 25. Kuratani M., Yanagisawa T., Ishii R., Matsuno M., Si S. Y., Katsura K., Ushikoshi-Nakayama R., Shibata R., Shirouzu M., Bessho Y., Yokoyama S. (2014) Crystal structure of tRNA m1A58 methyltransferase TrmI from Aquifex aeolicus in complex with S-adenosyl-l-methionine. J. Struct. Funct. Genomics 15, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guelorget A., Roovers M., Guérineau V., Barbey C., Li X., Golinelli-Pimpaneau B. (2010) Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase. Nucleic Acids Res. 38, 6206–6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guelorget A., Barraud P., Tisné C., Golinelli-Pimpaneau B. (2011) Structural comparison of tRNA m1A58 methyltransferases revealed different molecular strategies to maintain their oligomeric architecture under extreme conditions. BMC Struct. Biol. 11, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roovers M., Kaminska K. H., Tkaczuk K. L., Gigot D., Droogmans L., Bujnicki J. M. (2008) The YqfN protein of Bacillus subtilis is the tRNA: m1A22 methyltransferase (TrmK). Nucleic Acids Res. 36, 3252–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. (2012) A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583–11593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kempenaers M., Roovers M., Oudjama Y., Tkaczuk K. L., Bujnicki J. M., Droogmans L. (2010) New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res. 38, 6533–6543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chujo T., Suzuki T. (2012) Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA 18, 2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watanabe K., Shinma M., Oshima T., Nishimura S. (1976) Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 33. Horie N., Hara-Yokoyama M., Yokoyama S., Watanabe K., Kuchino Y., Nishimura S., Miyazawa T. (1985) Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24, 5711–5715 [DOI] [PubMed] [Google Scholar]
- 34. Shigi N., Suzuki T., Tamakoshi M., Oshima T., Watanabe K. (2002) Conserved bases in the TΨC loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J. Biol. Chem. 277, 39128–39135 [DOI] [PubMed] [Google Scholar]
- 35. Tomikawa C., Yokogawa T., Kanai T., Hori H. (2010) N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability through a tRNA modification network. Nucleic Acids Res. 38, 942–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takeda H., Toyooka T., Ikeuchi Y., Yokobori S., Okadome K., Takano F., Oshima T., Suzuki T., Endo Y., Hori H. (2006) The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells 11, 1353–1365 [DOI] [PubMed] [Google Scholar]
- 37. Shigi N., Suzuki T., Terada T., Shirouzu M., Yokoyama S., Watanabe K. (2006) Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281, 2104–2113 [DOI] [PubMed] [Google Scholar]
- 38. Hoseki J., Yano T., Koyama Y., Kuramitsu S., Kagamiyama H. (1999) Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 126, 951–956 [DOI] [PubMed] [Google Scholar]
- 39. Hoseki J., Okamoto A., Takada N., Suenaga A., Futatsugi N., Konagaya A., Taiji M., Yano T., Kuramitsu S., Kagamiyama H. (2003) Increased rigidity of domain structures enhances the stability of a mutant enzyme created by directed evolution. Biochemistry, 42, 14469–14475 [DOI] [PubMed] [Google Scholar]
- 40. Yokogawa T., Kitamura Y., Nakamura D., Ohno S., Nishikawa K. (2010) Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 38, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kazayama A., Yamagami R., Yokogawa T., Hori H. (2015) Improved solid-phase DNA probe method for tRNA purification: large scale preparation, and alteration of DNA fixation. J. Biochem., 10.1093/jb/mvu089 [DOI] [PubMed] [Google Scholar]
- 42. Grosjean H., Edqvist J., Stråby K. B., Giegé R. (1996) Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 255, 67–85 [DOI] [PubMed] [Google Scholar]
- 43. Constantinesco F., Motorin Y., Grosjean H. (1999) Transfer RNA modification enzymes from Pyrococcus furiosus: detection of the enzymatic activities in vitro. Nucleic Acids Res. 27, 1308–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., Kumagai I., Watanabe K. (1998) Substrate recognition of tRNA (guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 273, 25721–25727 [DOI] [PubMed] [Google Scholar]
- 45. Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250, 546–551 [DOI] [PubMed] [Google Scholar]
- 46. Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185, 435–440 [DOI] [PubMed] [Google Scholar]
- 47. Shi H, Moore P. B. (2000) The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gu X. R., Santi D. V. (1991) The T-arm of tRNA is a substrate for tRNA (m5U54)-methyltransferase. Biochemistry 30, 2999–3002 [DOI] [PubMed] [Google Scholar]
- 49. Okamoto H., Watanabe K., Ikeuchi Y., Suzuki T., Endo Y., Hori H. (2004) Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 279, 49151–49159 [DOI] [PubMed] [Google Scholar]
- 50. Matsumoto K., Toyooka T., Tomikawa C., Ochi A., Takano Y., Takayanagi N., Endo Y., Hori H. (2007) RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett. 581, 1599–1604 [DOI] [PubMed] [Google Scholar]
- 51. Yamagami R., Yamashita K., Nishimasu H., Tomikawa C., Ochi A., Iwashita C., Hirata A., Ishitani R., Nureki O., Hori H. (2012) The tRNA recognition mechanism of folate/FAD-dependent tRNA methyltransferase (TrmFO). J. Biol. Chem. 287, 42480–42494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alian A., Lee T. T., Griner S. L., Stroud R. M., Finer-Moore J. (2008) Structure of a TrmA-RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 105, 6876–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Awai T., Ochi A., Ihsanawati, Sengoku T., Hirata A., Bessho Y., Yokoyama S., Hori H. (2011) Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the x-ray crystal structure. J. Biol. Chem. 286, 35236–35246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ochi A., Makabe K., Yamagami R., Hirata A., Sakaguchi R., Hou Y. M., Watanabe K., Nureki O., Kuwajima K., Hori H. (2013) The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process. J. Biol. Chem. 288, 25562–25574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ishida K., Kunibayashi T., Tomikawa C., Ochi A., Kanai T., Hirata A., Iwashita C., Hori H. (2011) Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 39, 2304–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grawunder U., Schön A., Sprinzl M. (1992) Sequence and base modifications of two phenylalanine-tRNAs from Thermus thermophilus HB8. Nucleic Acids Res. 20, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grosjean H., Constantinesco F., Foiret D., Benachenhou N. (1995) A novel enzymatic pathway leading to 1-methylinosine modification in Haloferax volcanii tRNA. Nucleic Acids Res. 23, 4312–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Constantinesco F., Motorin Y., Grosjean H. (1999) Characterisation and enzymatic properties of tRNA(guanine 26, N2,N2)-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 291, 375–392 [DOI] [PubMed] [Google Scholar]
- 59. Awai T., Kimura S., Tomikawa C., Ochi A., Ihsanawati, Bessho Y., Yokoyama S., Ohno S., Nishikawa K., Yokogawa T., Suzuki T., Hori H. (2009) Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 284, 20467–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]