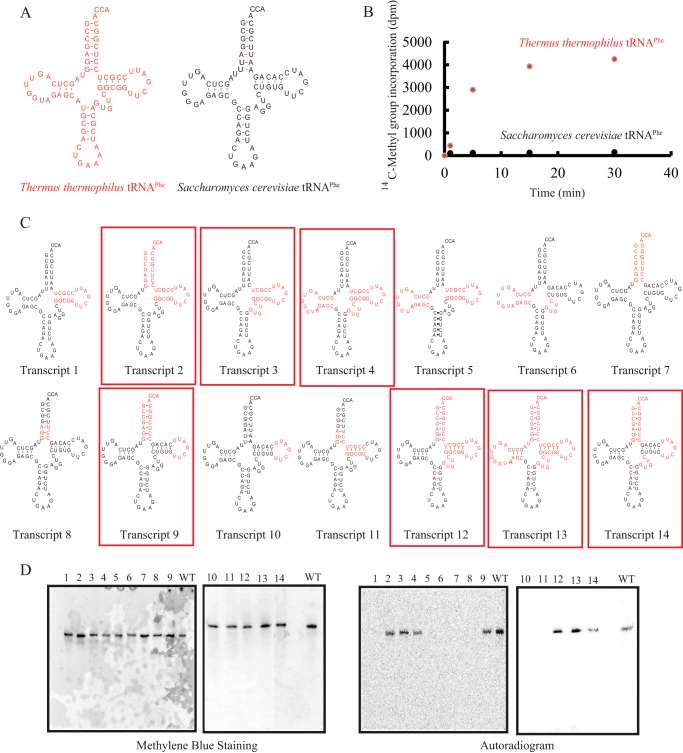
FIGURE 2.
Chimeric tRNA transcripts of T. thermophilus and S. cerevisiae tRNAPhe. A, T. thermophilus tRNAPhe (left) and S. cerevisiae (right) tRNAPhe are depicted as clover leaf structures. To distinguish the two tRNAs, T. thermophilus tRNAPhe is indicated in red. B, the methyl group acceptance activities of T. thermophilus and S. cerevisiae tRNAPhe were compared. S. cerevisiae tRNAPhe was methylated very slowly by TrmI: the initial velocity of methyl group acceptance of S. cerevisiae tRNAPhe was ∼4% of that of T. thermophilus tRNAPhe. C, 14 chimeric tRNA transcripts are depicted as clover leaf structures. The sequences derived from T. thermophilus tRNAPhe are colored in red. The chimeric transcripts whose methyl group acceptance activities were more than 50% of that of T. thermophilus tRNAPhe are enclosed with red boxes. D, after methylation by TrmI and [methyl-14C]AdoMet, the chimeric tRNA transcripts (0.02 A260 unit each) were analyzed by 10% PAGE (7 m urea) and then visualized by methylene blue staining (left panels). The lane numbers correspond to the transcript numbers shown in C. WT indicates the T. thermophilus wild-type tRNAPhe transcript. To analyze the methyl group acceptance activities of the tRNA transcripts, the same gels were subjected to autoradiography (right panels). The kinetic parameters are summarized in Table 1.