FIG. 6.
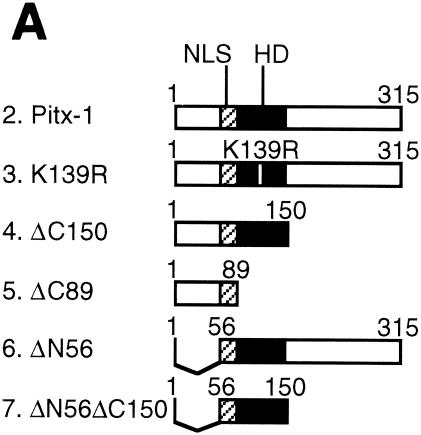
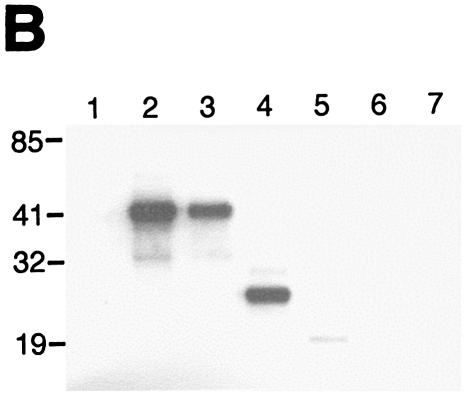
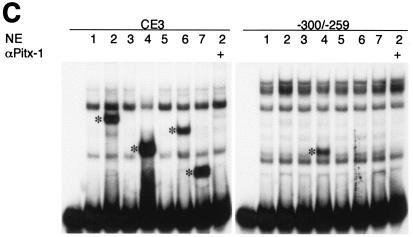
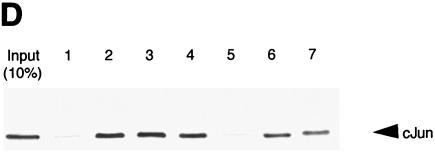
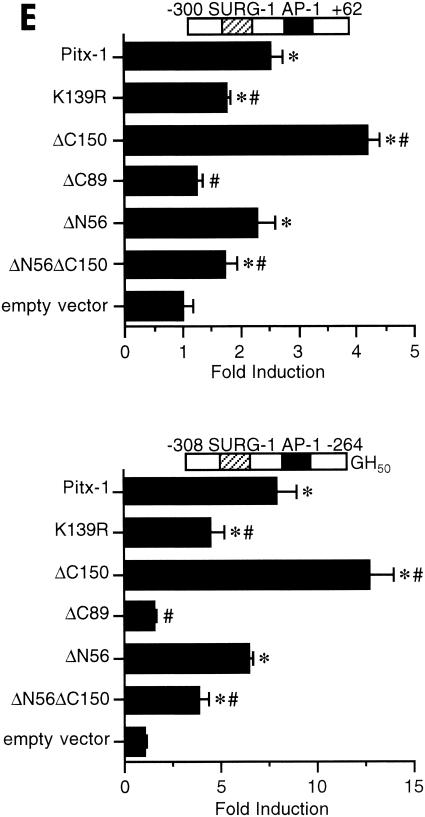
The Pitx-1 HD is necessary for transactivation of the mGnRHR gene promoter. (A) Schematic illustration of structures for intact Pitx-1 and the mutant Pitx-1 constructs generated. One set of constructs was generated in the pcDNA3 expression vector, and another set was generated in the pGEX-4T-2 expression vector for GST fusion proteins. NLS, nuclear localization signal. (B) Western blot analysis was performed using nuclear extracts (10 μg/lane) of 293T cells transfected with each Pitx-1 construct. Lanes: 1, empty pcDNA3 vector; 2 to 7, Pitx-1 constructs corresponding to the structures illustrated in panel A. Molecular masses (in kilodaltons) of protein markers are indicated on the left side of the gel. (C) EMSA analyses were performed using nuclear extracts (20 μg/lane) of 293T cells transfected with each Pitx-1 construct. CE3 or −300/−259 was used as probe. Numbers for each lane denote the Pitx-1 constructs as indicated for panels A and B. In the last lane of each gel, α-Pitx-1 antibody was added to the EMSA reaction mixture with nuclear extract containing intact Pitx-1. NE, nuclear extract; *, Pitx-1-DNA complexes. (D) GST pull-down analysis using hc-Jun (0.1 fpu/lane) and intact or mutant GST/Pitx-1 proteins (1 μg/lane) was performed as described in Materials and Methods. Lanes: 1, GST; 2 to 7, GST/Pitx-1 protein constructs corresponding to the structures illustrated in panel A. For the 10% input lane, 0.01 fpu of hc-Jun was loaded. (E) Luc reporter genes fused to −300/+62 or −308/−264/GH50 were cotransfected into CV-1 cells with either intact or mutant Pitx-1 expression vectors, or with empty vector, pcDNA3. Data are plotted as the induction by Pitx-1 over the empty expression vector control response for each reporter. Data are presented as the mean ± SEM of three independent experiments. *, P < 0.05 versus pcDNA3 empty vector; #, P < 0.05 versus intact Pitx-1.