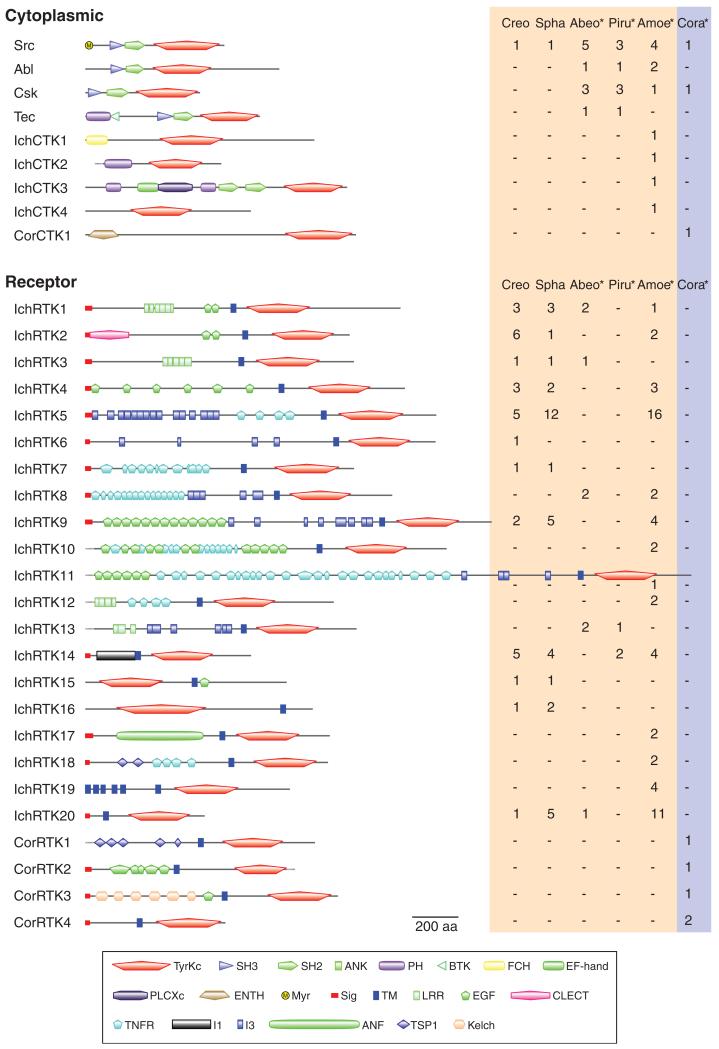
FIG. 2.
Ichthyosporean and corallochytrean TKs. TKs found in the whole genome data of two ichthyosporeans (Creo, Creolimax; Spha, Sphaeroforma) and the RNAseq data (asterisks) from three other ichthyosporeans (Abeo, Abeoforma; Piru, Pirum; Amoe, Amoebidium), and a corallochytrean (Cora, Corallochytrium) are classified according to their domain architectures and phylogenetic positions. CTKs are classified into ichthyosporean-specific families (IchCTK1-4) or corallochytrean-specific families (CorCTK1) unless they are homologous to metazoan CTKs. The domain architecture of a family member is schematically shown. The number of genes within each family is shown on the right (red and blue shades for ichthyosporeans and corallochytreans, respectively). See supplementary figure S1, Supplementary Material online, for the full list of annotated TKs. ANF, atrial natriuretic factor receptor-like ligand binding region; ANK, ankyrin repeats; BTK, Bruton’s tyrosine kinase Cys-rich motif; CLECT, C-type lectin (CTL) or carbohydrate-recognition domain (CRD); EF-hand, EF-hand-like domain; EGF, epidermal growth factor-like domain; ENTH, epsin N-terminal homology domain; FCH, Fes/CIP4 homology domain; I1, I1 domain; I3, I3 domain; Kelch, Kelch motif; LRR, leucine-rich repeat; Myr, predicted myristoylation site; PH, Pleckstrin homology domain; PLCXc, phospholipase C catalytic domain X; SH2, Src homology 2 domain; SH3, Src homology 3 domain; Sig, signal peptide; TM, transmembrane segment; TNFR, tumor necrosis factor receptor/nerve growth factor receptor repeat; TSP1, thrombospondin type 1 repeats; TyrKc, tyrosine kinase catalytic domain. Incomplete sequences are shown by dotted lines.