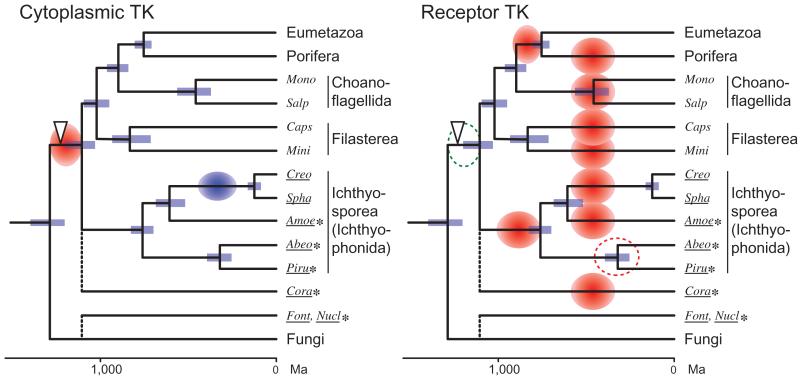
FIG. 6.
Evolution of the holozoan TK diversity. The history of family diversification of the metazoan-type (group A) TK is schematically represented. The tree is based on the time-calibrated phylogeny by Bayesian inference. Blue bars represent the 95% range of highest probability density (Drummond et al. 2012) of the node age. Red and blue circles represent extensive TK diversification and massive reduction of TK diversity, respectively. The period of RTK diversification in Abeoforma whisleri and Pirum gemmata remains unclear (red dotted circle), because RNAseq data may represent a minor fraction of TK diversity. The open arrowhead represents the evolution of the complex pTyr-mediated signaling system with the diversified pTyr signaling modules, as well as their enhanced combination. Note that we do not exclude the possibility of basal holozoan expansion of group A RTKs (green dotted circle), which may be obscured today by their rapid turnover during the holozoan evolution. Amoe, Amoebidium; Abeo, Abeoforma; Creo, Creolimax; Cora, Corallochytrium; Caps, Capsaspora; Font, Fonticula; Mini, Ministeria; Mono, Monosiga brevicollis; Nucl., Nuclearia; Piru, Pirum; Spha, Sphaeroforma; Salp, Salpingoeca. Species that were newly analyzed in this study are underlined, with the RNAseq data labeled with asterisks.