Abstract
Understanding the origin and evolution of the eukaryotic cell and the full diversity of eukaryotes is relevant to many biological disciplines. However, our current understanding of eukaryotic genomes is extremely biased, leading to a skewed view of eukaryotic biology. We argue that a phylogeny-driven initiative to cover the full eukaryotic diversity is needed to overcome this bias. We encourage the community: (i) to sequence a representative of the neglected groups available at public culture collections, (ii) to increase our culturing efforts, and (iii) to embrace single cell genomics to access organisms refractory to propagation in culture. We hope that the community will welcome this proposal, explore the approaches suggested, and join efforts to sequence the full diversity of eukaryotes.
Keywords: eukaryotic genomics, phylogeny, ecology, eukaryotic tree of life, culture collections, culturing bias, single cell genomics
The need for a phylogeny-driven eukaryotic genome project
Eukaryotes are the most complex of the three domains of life. The origin of eukaryotic cells and their complexity remains one of the longest-debated questions in biology, famously referred to by Roger Stanier as the ‘greatest single evolutionary discontinuity’ in life [1]. Thus, understanding how this complex cell originated and how it evolved into the diversity of forms we see today is relevant to all biological disciplines including cell biology, evolutionary biology, ecology, genetics, and biomedical research. Progress in this area relies heavily on both genome data from extant organisms and on an understanding of their phylogenetic relationships.
Genome sequencing is a powerful tool that helps us to understand the complexity of eukaryotes and their evolutionary history. However, there is a significant bias in eukaryotic genomics that impoverishes our understanding of the diversity of eukaryotes, and leads to skewed views of what eukaryotes even are, as well as their role in the environment. This bias is simple and widely recognized: most genomics focuses on multicellular eukaryotes and their parasites. The problem is not exclusive to eukaryotes. The launching of the so-called ‘Genomic Encyclopedia of Bacteria and Archaea’ [2] has begun to reverse a similar bias within prokaryotes, but there is currently no equivalent for eukaryotes. Targeted efforts have recently been initiated to increase the breadth of our genomic knowledge for several specific eukaryotic groups, but again these tend to focus on animals [3], plants [4], fungi [5], their parasites [6], or opisthokont relatives of animals and fungi [7]. Unfortunately, a phylogeny-driven initiative to sequence eukaryotic genomes specifically to cover the breadth of their diversity is lacking. The tools already exist to overcome these biases and fill in the eukaryotic tree, and we therefore hope that researchers will be inspired to explore these tools and embrace the prospect of working towards a community-driven initiative to sequence the full diversity of eukaryotes.
The multicellular effect
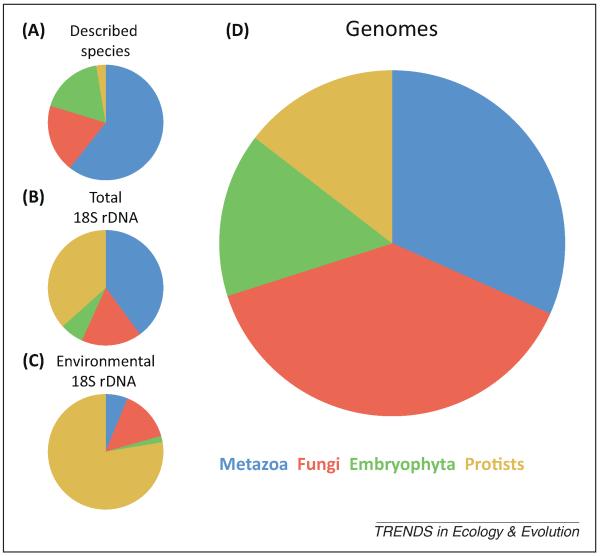
It is not surprising that the first and main bias in the study of eukaryotes arises from our anthropocentric view of life. More than 96% of the described eukaryotic species are either Metazoa (animals), Fungi, or Embryophyta (land plants) [8] (Figure 1A) – which we call the ‘big three’ of multicellular organisms (even though the Fungi also include unicellular members such as the yeasts). However, these lineages only represent 62% of the 18S rDNA (see Glossary) Genbank sequences (Figure 1B), which is of course a biased sample, or 23% of all operational taxonomic units (OTUs) in environmental surveys (Figure 1C). This bias is not new; research has historically focused on these three paradigmatic eukaryotic kingdoms, which are indeed important, but are also simply more conspicuous and familiar to us. In genomics this bias is amplified considerably: 85% of the completed or projected genome projects {as shown by the Genomes On-Line Database (GOLD) [9]} belong to the ‘big three’ (Figure 1D). Moreover, even within these groups there are biases. For example, many diverse invertebrate groups suffer from a lack of genomic data as keenly as do microbial groups. This makes for a pitiful future if we aim to understand and appreciate the complete eukaryotic tree of life. If we do not change this trend we risk neglecting the majority of eukaryotic diversity in future genomic or metagenomic-based ecological and evolutionary studies. This would provide us with a far from realistic picture.
Figure 1.
Relative representation of metazoans, fungi, and land plants versus all the other eukaryotes in different databases. (A) Relative numbers of described species according to the CBOL ProWG (n = 2 001 573). (B) Relative numbers of 18S rDNA OTU97 in GenBank (n = 22 475). (C) Relative number of environmental 18S rDNA OTU97 in GenBank (n = 1165). (D) Relative number of species with a genome project completed or in progress according to GOLD, per eukaryotic group (n = 1758). Data in panels A–C are from [8]. Abbreviations: CBOL ProWG, Consortium for the Barcode of Life Protist Working Group; GOLD, Genomes OnLine Database; OTU97, operational taxonomic unit (>97% sequence identity).
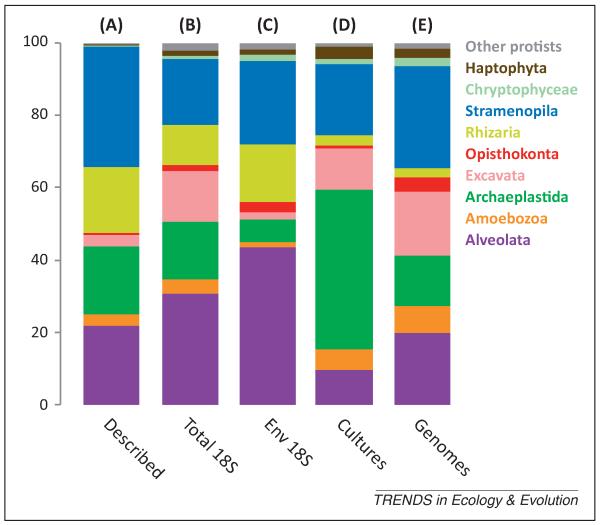
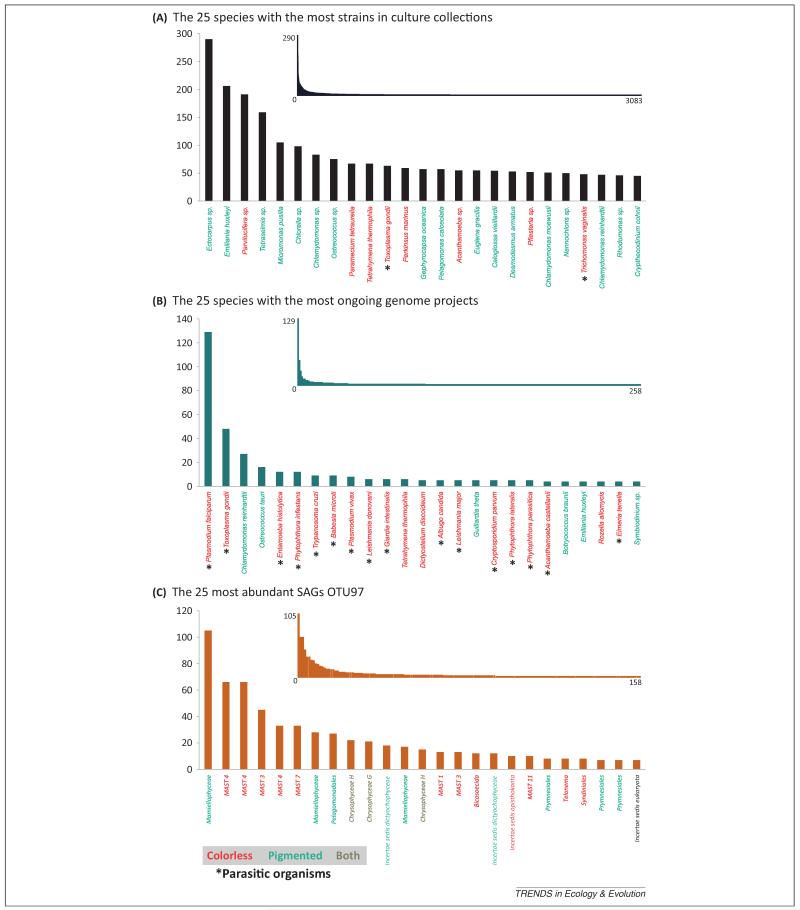
The ‘multicellular bias’ is the most serious, but is not alone. The eukaryotic groups with most species deposited in culture collections and/or genome projects are also biased towards either those containing mainly phototrophic species or those that are parasitic and/or economically important (Figure 2). For example, both Archaeplastida and Stramenopila have more cultured species than other eukaryotes as a result of a long phycological tradition and the well-provided phycological culture collections [10], and also because they are easier to maintain in culture than heterotrophs. In both cases this translates to a comparatively large number of genome projects: several genomic studies target photosynthetic stramenopiles[11,12] and, owing to their economic relevance in the agriculture, the peronosporomycetes [13]. In addition, the apicomplexans within the Alveolata are also relatively well studied at the genomic level because they contain important human and animal parasites [14] such as Plasmodium and Toxoplasma. If we look instead at the number of sequenced strains rather than species, these biases are increased further (Figure 3). As a result, a significant proportion of the retrieved cultures and genomes correspond to different strains of the same dominant species. Therefore, we have a pool of species that have been redundantly cultured and sequenced.
Figure 2.
Relative representation of eukaryotic supergroup diversity in different databases. (excluding metazoans, fungi, and land plants). (A) Percentage of described species per eukaryotic supergroup according to the CBOL ProWG. (B) Percentage of 18S rDNA OTU97 per eukaryotic supergroups in GenBank. (C) Percentage of environmental 18S rDNA OTU97 per eukaryotic supergroups. (D) Percentage of species with a cultured strain in any of the analyzed culture collections. Culture data are from five large protist culture collections (n = 3084) (the American Type Culture Collection, Culture Collection of Algae and Protozoa [24], the Roscoff Culture Collection [25], the National Center for Marine Algae and Microbiota [26] and the Culture Collection of Algae at Göttingen University [27]). (E) Relative numbers of species with a genome project completed or in progress according to GOLD, per eukaryotic group. Data from panels A–C are from [8]. Data from panels D and E are publicly available and the taxonomic analysis can be found in the supplementary data online. Abbreviations: CBOL ProWG, Consortium for the Barcode of Life Protist Working Group; Env 18S, environmental 18S rDNA sequences; GOLD, Genomes OnLine Database; OTU97, operational taxonomic unit (>97% sequence identity).
Figure 3.
Eukaryotic a diversity distribution among the analyzed databases. (A) The 25 species with the most strains represented in the analyzed culture collections. (B) The 25 speciesa with the most ongoing genome projects. (C) The 25 most abundant SAGs OTU97 in the analyzed dataset. Abbreviations: MAST, marine stramenopile; OTU97, operational taxonomic unit (>97% sequence identity); SAG, single amplified genome.
aSome strains are not described at the species level and have been grouped by genus. Therefore they may represent more than a single species.
The missing branches of the eukaryotic tree of life
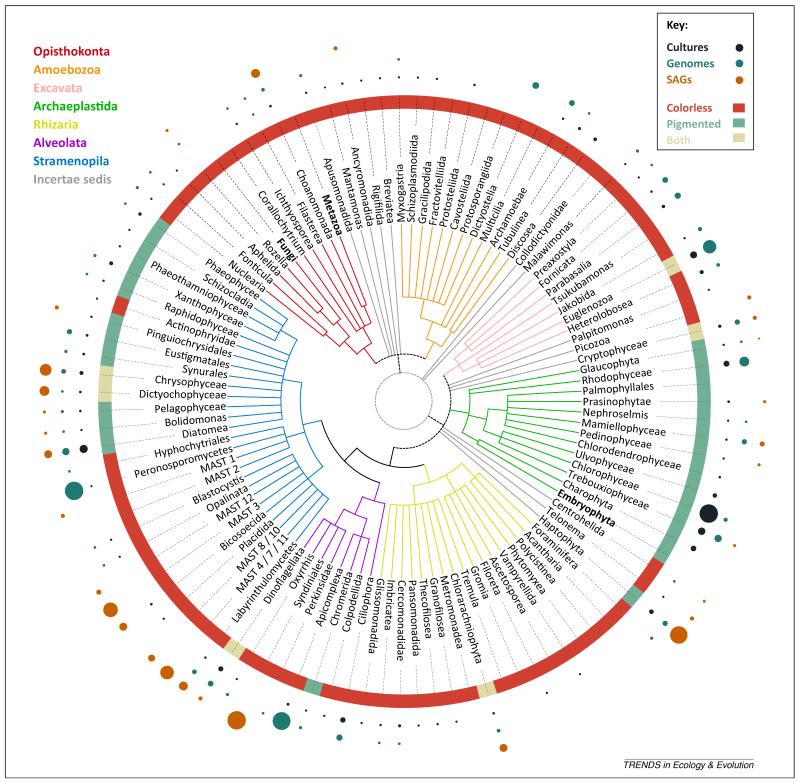
Although we lack an incontrovertible, detailed phylogenetic tree of the eukaryotes, a consensus tree is emerging thanks to molecular phylogenies [15]. The five monophyletic supergroups of eukaryotes are summarized in Box 1. The distribution of cultured and sequenced species over the tree provides a broad overview of our current knowledge of eukaryotic diversity (Figure 4). However, a quarter of the represented lineages lack even a single culture in any of the analyzed culture collections and, notably, 51% of them lack a genome. The most important gaps are within the Rhizaria, the Amoebozoa, and the Stramenopila, where many lineages are still underrepresented. However, many other lineages that lack any representative genome sequence are also found in the relatively well-described Opisthokonta and Excavata groups. This map is likely to be incomplete because several genome projects may not be reflected in the GOLD database, and because many cultures are not deposited in culture collections, but the overall trends probably afford an accurate representation of the biases we currently face.
Box 1. The five eukaryotic supergroups.
Thanks to molecular phylogenetics, to ultrastructural analyses, and to the efforts of many researchers, we have in recent years advanced significantly our understanding of the tree of eukaryotes. According to the most recent consensus taxonomy [28], the eukaryotes can be divided into five monophyletic supergroups. We here introduce these supergroups, detailing some specific features of each.
Amoebozoa: this group consists of amoeboid organisms, most of them possessing a relatively simple life cycle and limited morphological features, as well as a few flagellated organisms [30]. They are common free-living protists inhabiting marine, freshwater, and terrestrial environments. Some well-known amoebozoans include the causative agent of amoebiasis (Entamoeba histolytica) and Dictyostelium sp., a model organism used in the study of the origin of multicellularity.
Archaeplastida: also known as ‘the green lineage’ or Viridiplantae, this group comprises the green algae and the land plants. The Archaeplastida is one of the major groups of oxygenic photosynthetic eukaryotes [31]. Green algae are diverse and ubiquitous in aquatic habitats. The land plants are probably the most dominant primary producers on terrestrial ecosystems. Both green algae and land plants have historically played a central role in the global ecosystem.
Excavata: the group Excavata was proposed based of shared morphological characters [32], and was later confirmed through phylogenomic analyses [33]. Most members of this group are heterotrophic organisms, among them some well-known human parasites such as Trichomonas vaginalis (the agent of trichomoniasis) and Giardia lamblia (the agent of giardiasis), as well as animal parasites such as Leishmania sp. (the agent of leishmaniasis) as well as Trypanosoma brucei, and Trypanosoma cruzi (the agents of sleeping sickness and Chagas disease respectively).
Opisthokonta: the opisthokonts include two of the best-studied kingdoms of life: the Metazoa (animals) and the Fungi. Recent phylogenetic and phylogenomic analyses have shown that the Opisthokonta also include several unicellular lineages [34]. These include the Choanoflagellata (the closest unicellular relatives of the animals) and the Ichthyospora (that include several fish parasites that impact negatively on aquaculture).
SAR (Stramenopila – Alveolata, and Rhizaria): three groups that have been historically studied separately. Phylogenetic analyses, however, have shown that those three groups share a common ancestor, forming a supergroup known as SAR [36]. This eukaryotic assemblage comprises the highest diversity within the protists.
Stramenopila: also known as heterokonts, the stramenopiles include a wide range of ubiquitous phototrophic and heterotrophic organisms [37]. Most are unicellular flagellates but there are also some multicellular organisms, such as the giant kelps. Other relevant members of the Stramenopila are the diatoms (algae contained within a silica cell wall), the chrysophytes (abundant in freshwater environments), the MAST (marine stramenopile) groups (the most abundant microbial predators of the ocean), and plant parasites such as the Peronosporomycetes.
Alveolata: a widespread group of unicellular eukaryotes that have adopted diverse life strategies such as predation, photoautotrophy, and intracellular parasitism [29]. They include some environmentally relevant groups such as the Syndiniales, the Dinoflagellata, and the ciliates (Ciliophora), as well as the Apicomplexa group that contains notorious parasites such as Plasmodium sp. (the agent of malaria), Toxoplasma sp. (the agent of toxoplasmosis), and Cryptosporidium sp.
Rhizaria: this is a diverse group of mostly heterotrophic unicellular eukaryotes including both amoeboid and flagellate forms [35]. Two iconic protist groups, Haeckel’s Radiolaria and the Foraminifera, are members of the Rhizaria. Foraminifera have been very useful in paleoclimatology and paleoceanography due to their external shell that can be detected in the fossil record.
Incertae sedis: Latin for ‘of uncertain placement’, a term used to indicate those organisms or lineages with unclear taxonomical position.
Figure 4.
The tree of eukaryotes, showing the distribution of current effort on culturing, genomics, and environmental single amplified genome (SAG) genomics for the main protistan lineages. Eukaryotic schematic tree representing major lineages. Colored branches represent the seven main eukaryotic supergroups, whereas grey branches are phylogenetically contentious taxa. The sizes of the dots indicate the proportion of species/OTU97 in each database. Culture data are from the analyzed publicly available protist culture collections (n = 3084). Genome data were extracted from the Genomes OnLine Database (GOLD) (n = 258) [9]. SAGs of OTU97 correspond to those retrieved during the Tara Oceans cruise (n = 158) (M.E.S., unpublished data). Taxonomic annotation of all datasets is based on [28]. The ‘big three’ (in bold) have been excluded from this analysis. Abbreviation: OTU97, operational taxonomic unit (>97% sequence identity).
Filling the gaps: how to
Although there may not be bad choices when selecting organisms for genome sequencing, there are certainly better choices if we aim to understand eukaryotic diversity. We argue that at least some of the effort should be specifically directed towards filling the gaps in the eukaryotic tree of life, focusing on those lineages that occupy key phylogenetic positions. How can that be done? One option is to sequence more cultured organisms. In fact, 95% of protist species in culture are not yet targeted for a genome project (Figure S1 in the supplementary data online). Thus, by obtaining the genome of some available cultured lineages that have not yet been sequenced, we could easily fill some of the important gaps of the tree, including some heterotrophic Stramenopila, Amoebozoa, and Rhizaria. However, selecting species that are available in culture is itself strongly biasing, and most lineages remain without any cultured representative [16]. Publicly accessible protist collections [such as the American Type Culture Collection (ATCC) and the Culture Collection of Algae and Protozoa (CCAP); summarized in Box 2] are considerably smaller than their bacterial or fungal counterparts. Among the reasons is the lack of a required, systematic deposit of newly described taxa, in contrast to the situation for bacteria [17]. Notably, and unfortunately, half of the species with genome projects completed or in progress are not deposited in any of the five analyzed publicly accessible culture collections. To avoid more ‘lost cultures’ in the future the community should establish and adopt standard procedures similar to those used in bacteriology to release cultures to protist collections. The whole community will benefit from this in the short and long term. In addition, there is an inherent technical bias in culturing, as well as a bias in culturing efforts. For example, phototrophic representatives of Stramenopila and Alveolata tend to have more cultures available than their heterotrophic counterparts (Figure 4). Indeed, 70.6% of the most common protist strains present in culture collections are phototrophic organisms (Figure 3). Therefore there is a need both to increase the culturing effort for a wider variety of environments and to develop novel and alternative culture techniques to retrieve refractory organisms [18], both of which take time, energy, and funding. Importantly, culture collections will need to be supported so that they can take on the challenge of maintaining more cultures and open their scope to include more difficult organisms that tend to be excluded from existing collections, in particular heterotrophs.
Box 2. Protist culture collections.
Culture collections are cornerstones for the development of all microbiological disciplines. Cultures are key to the establishment of model organisms and, therefore, to a better understanding of their biology. Below we describe some of the major protistan collections.
ATCC (American Type Culture Collection; Manassas, Virginia, USA): a private, non-profit biological resource center established in 1925 with the aim of creating a central collection to supply microorganisms to scientists all over the world (http://www.atcc.org). ATCC collections include a great variety of biological materials such as cell lines, molecular genomics tools, microorganisms, and bioproducts. The microorganism collection includes more than 18 000 strains of bacteria, 3000 different types of viruses, over 49 000 yeast and fungal strains, and 2000 strains of protists.
CCAP (Culture Collection of Algae and Protozoa; Oban, Scotland, UK): a culture collection funded by the UK Natural Environmental Research Centre (NERC) that contains algae and protozoa from both freshwater and marine environments. The foundations of CCAP (http://www.ccap.ac.uk) were laid by Prof. Ernst Georg Pringsheim and his collaborators and the cultures they established at the Botanical Institute of the German University of Prague in the 1920s. Pringsheim moved to England where the collection was expanded and taken over by Cambridge University in 1947. In 1970 these cultures formed the basis of the Culture Centre of Algae and Protozoa that later became the modern CCAP.
NCMA (Provasoli-Guillard National Center for Marine Algae and Microbiota, East Boothbay, Maine, USA): this integrated collection of marine algae, protozoa, bacteria, archaea, and viruses was named a National Center and Facility by the US Congress in 1992. The NCMA (http://ncma.bigelow.org) originated from private culture collections established by Dr Luigi Provasoli at Yale University and Dr Robert R.L. Guillard at Woods Hole Oceanographic Institution. When it was born in the 1980s it was known as the Culture Collection of Marine Phytoplankton (CCMP) and provided to the community algal cultures of scientific interest or for aquaculture.
RCC (Roscoff Culture Collection; Roscoff, France): this collection (http://www.roscoff-culture-collection.org) is located at the Station Biologique de Roscoff and is closely linked to the Oceanic Plankton group of this institution. They maintain more than 3000 strains of marine phytoplankton, especially picoplankton and picoeukaryotes from various oceanic regions. Most of the strains are available for distribution whereas others are in the process of being described.
SAG (Sammlung von Algenkulturen: Culture Collection of Algae at Göttingen University, Göttingen, Germany): the SAG is a non-profit organization maintained by the University of Göttingen (http://www.epsag.uni-goettingen.de). The collection primarily contains microscopic algae and cyanobacteria from freshwater or terrestrial habitats, but there are also some marine algae. With more than 2400 strains, the SAG is among the three largest culture collections of algae in the world. Prof. Pringsheim is also the founder of the SAG: it was initiated in 1953 when he returned to Göttingen after his time as a refugee scientist in England. From then on the Pringsheim algal collection has been growing and evolving into the service collection we know nowadays.
A complementary option to increase the breadth of eukaryotic genomics is to use single cell genomics (SCG) [19]. Although the technology is still developing, this is probably the best way we have today to retrieve genomic information from abundant microbial eukaryotes that are ecologically relevant but are refractory to being cultured. For example, the single amplified genomes (SAGs) from different global oceanic sites obtained during the Tara Oceans cruise (M.E.S., unpublished data) fill reasonably well the culture and genomic gaps that some of the most abundant groups in the oceans suffer from (Figure 4). In particular, a significant fraction of the SAGs correspond to uncultured organisms such as the marine stramenopiles MAST-4 and MAST-7 [20], chrysophyte groups H and G [21], and the Syndiniales [22]. Importantly, sequence tagging shows that only 10% of the SAGs are present in any culture collection, and only 2.5% have an ongoing genome project (based on cultured taxa). It is worth mentioning that the SAGs so far available represent only marine microeukaryotes. Thus, although the analyzed SAGs certainly overcome part of the bias, they do not cover the full diversity of eukaryotes.
Given the potential of SAGs to improve further our understanding of eukaryotic diversity, an important question to ask is whether high-quality genome data can be acquired from SAGs [19]. Currently, there seems to be a diversity of outcomes when using SAGs owing to the bias introduced by the whole-genome amplification procedure. The completeness range of the retrieved genome varies from less than 10% to a complete genome, and depends on the intrinsic properties of the cell studied as well as on the amplification method [23]. Culture certainly provides a more reliable way to obtain a genome of high quality at present, and a species in culture also provides researchers with a direct window to the biology of the organism and post-genomic research. Auto-ecological experiments, ultrastructure analyses, and even functional experiments can all be performed in culture, thereby providing a deeper context for the genome and the organism. However, in light of the lack of data we currently face, and the unlikelihood that a significant increase in resources for cultivation will soon appear, we argue strongly that genomic sequencing of SAGs is an important complement to culture-based research in furthering our understanding of eukaryotic diversity.
Make the tree thrive: a call to action
Genome sequences have cast invaluable light on the classification of organisms, notably in many cases where particular species were misclassified (Box 3). However, the available genome sequences of eukaryotes do not inform us only about the biology of the particular organism. They also make significant contributions to our understanding of eukaryotic biology in general, and to large-scale evolutionary and ecological processes. Nevertheless, for this potential to be completely fulfilled we must sample broadly, and there are currently important gaps in the diversity of eukaryotic genome sequences that undermine our efforts to capitalize on this potential. Understanding the whole of eukaryotic diversity will doubtless contribute to our understanding of specific biological questions, including some of our more pernicious problems in medicine, agriculture, evolution, and ecology.
Box 3. The rectification of names and our understanding of eukaryotic biology.
A better understanding on the eukaryotic diversity has a deep impact in several biological disciplines such as medicine, agriculture, evolution, and ecology. A large body of research backs up this statement. Below we mention a few examples that illustrate the power of having a better understanding of the diversity, biology, and evolution of eukaryotes.
Medicine has greatly benefited from evolutionary studies in eukaryotes. Studies on the genome and biology of close relatives of parasites have provided unique insights into analogous molecular mechanisms involved in the clinical effects of parasites. A proper taxonomic assignment of pathogenic organisms has also been the key to fighting them. A good example is Pneumocystis, an opportunistic pathogen affecting immunocompromised patients, predominantly HIV-infected. Pneumocystis was considered for years to a protozoan of unclear taxonomic assignment. It was not until molecular data allowed researchers to properly assign Pneumocystis to the fungi, in 1988, that adequate treatments based on antifungal agents could be used [38]. The opposite situation happened with the fungus-like Phytophthora, the causative agent of the potato blight. New molecular data showed that Phytophthora are peronosporomycetes (stramenopiles) within the order Peronosporales, and not fungi as previously thought, thus explaining the ineffective use of fungicides [39]. Further knowledge of its genome provided insights not only into its evolution but also into the potential reasons for its speed to form resistant forms [40].
It is, however, in evolutionary studies where the impact of having a broad taxon sampling of eukaryotes is more apparent. Indeed, and looking back in time, it is clear that the absence of key taxa in evolutionary analyses led to hypotheses that are now known to be in error. The fact is that to elucidate which genomic or morphological features have been conserved, which were ancestral to eukaryotes, and which are novel, one needs to perform comparative analyses that must include key taxa from each major eukaryotic lineage. For example: instances of lineage-specific gene loss in Choanoflagellatea and Fungi, and the absence of representative taxa from non-parasites Excavata and Rhizaria, confounded attempts to reconstruct accurately the gene content of the last unicellular ancestor of metazoans [41] and the last eukaryotic common ancestor [42], respectively.
Ecology is also influenced by a better understanding of eukaryote biology. The global ecological cycles are deeply influenced by several groups of eukaryotes, most of them unicellular. We have a good understanding of phototrophic eukaryotes that, together with the Cyanobacteria, drive most of the carbon cycle and the oxygen production on earth. Nevertheless, our understanding of heterotrophic protists remains insufficient. For example, both MASTs and the Syndiniales are extremely abundant in the oceans [43]. Therefore, they are surely influential in global processes. However, we cannot understand their role if we lack information on their metabolic pathways or biology, something we can only obtain from genomic data.
We propose that filling in the eukaryotic tree at the genomic level based on phylogenetic diversity should be a priority for the community. We also argue that this can be achieved by a combination of three complementary approaches. First, at least one genome from underrepresented lineages from which cultures are available should be sequenced. This is a straightforward problem, requiring phycologists, protistologists, culture collection curators, and genomic sequencing centers to coordinate efforts and expertise to choose the best target taxa and sequencing strategies. Second, efforts to culture diverse organisms should be supported, by sampling additional areas of the planet, developing novel techniques to include more recalcitrant species (especially heterotrophs), and by rewarding this difficult but essential task, especially in younger researchers before they conclude en masse that such crucial work is a professional dead-end. Such efforts are timeconsuming and have a built-in failure rate that makes them risky, and therefore policy changes will be helpful in order that funding agencies, universities, and research centers recognize the value of such work independently of the publication outcome. Finally, microbial ecologists and genomic centers should embrace the use of SCG and continue to improve the technology, which we believe will be the key to filling in missing parts of the tree in the short term. To coordinate all these efforts, funding agencies should also support the development of community resources such as publicly accessible culture collections and the maintenance of key taxa that are difficult to keep.
We believe strongly that the time is ripe to reverse the genome sequencing bias in the tree of eukaryotes. We now have in our hands all the elements needed to change this skewed view and further our understanding of eukaryotic biology and evolution. All that needs to change is the will and a joint coordinated initiative. Thus, we hope that the eukaryotic community will welcome this proposal to build a representative and diverse ‘Genomic Encyclopedia of Eukaryotes’ and collaborate to make this happen.
Supplementary Material
Glossary
- 18S rDNA
genes encoding the RNA of the small ribosomal subunit are found in all eukaryotes in many copies per genome. They are also highly expressed and its nucleotide structure combine well-conserved and variable regions. Because of these characteristics 18S rDNA has been used as a marker to identify and barcode eukaryotes at the species or genus level (with some exceptions). It is also the most widely used eukaryotic phylogenetic marker.
- Culturing bias
cultured microbial strains do not necessarily represent, and usually are not, the dominant members of the environment from which they were isolated. This bias affects bacteria, viruses, and protists. The culturing bias can be the result of a lack of continuous culturing efforts, or inadequate isolation and/or culturing strategies – or because, for whatever reason, some species in the environment may be refractory to isolation and culturing.
- Genomes OnLine Database (GOLD)
an online resource for comprehensive access to information regarding genome and metagenome sequencing projects, and their associated metadata (http://www.genomesonline.org/).
- Operational taxonomic unit (OTU)
an operational definition of a species or group of species. In microbial ecology, and in particular protist ecology, this operational definition is generally based in a percentage similarity threshold of the 18S rDNA (e.g., OTU97 refers to a cluster of sequences with >97% similarity that are inferred to represent a single taxonomic unit).
- Single amplified genomes (SAGs)
the products of single cell whole-genome amplification that can be further analyzed in similar ways to DNA extracts from pure cultures.
- Single cell genomics (SCG)
a method to amplify and sequence the genome of a single cell. The method consists of an integrated pipeline that starts with the collection and preservation of environmental samples, followed by physical separation, lysis, and whole-genome amplification from individual cells. This is followed by sequencing of the resulting material. SCG is a powerful complement to culture-based and environmental microbiology approaches [23].
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tree.2014.03.006.
References
- 1.Stanier RY, et al. The Microbial World. Prentice-Hall; 1957. [Google Scholar]
- 2.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennisi E. No genome left behind. Science. 2009;326:794–795. doi: 10.1126/science.326_794. [DOI] [PubMed] [Google Scholar]
- 4.Bennetzen J, Kellogg E. A plant genome initiative. Plant Cell. 1998;10:488–494. [Google Scholar]
- 5.Galagan JE, et al. Genomics of the fungal kingdom: insights into eukaryotic biology. Genome Res. 2005;15:1620–1631. doi: 10.1101/gr.3767105. [DOI] [PubMed] [Google Scholar]
- 6.Degrave WM, et al. Parasite genome initiatives. Int. J. Parasitol. 2001;31:532–536. doi: 10.1016/s0020-7519(01)00144-8. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Trillo I, et al. A phylogenomic investigation into the origin of metazoa. Mol. Biol. Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 8.Pawlowski J, et al. CBOL Protist Working Group, barcoding eukaryotic richness beyond the Animal, Plant, and Fungal Kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagani I, et al. The Genomes OnLine Database (GOLD) v.4, status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day JG, et al. Pringsheim’s living legacy: CCALA, CCAP, SAG and UTEX culture collections of algae. Nova Hedwigia. 2004;79:27–37. [Google Scholar]
- 11.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 12.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 13.Pais M, et al. From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol. 2013;14:211. doi: 10.1186/gb-2013-14-6-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Dooren GG, Striepen B. The algal past and parasite present of the apicoplast. Annu. Rev. Microbiol. 2013;67:271–289. doi: 10.1146/annurev-micro-092412-155741. [DOI] [PubMed] [Google Scholar]
- 15.He D, et al. An alternative root for the eukaryote tree of life. Curr. Biol. 2014;24:465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 16.del Campo J, et al. Culturing bias in marine heterotrophic flagellates analyzed through seawater enrichment incubations. Microb. Ecol. 2013;66:489–499. doi: 10.1007/s00248-013-0251-y. [DOI] [PubMed] [Google Scholar]
- 17.Lapage SP, et al. International Code of Nomenclature of Bacteria. ASM Press; 1992. http://www.ncbi.nlm.nih.gov/books/NBK8817/ [PubMed] [Google Scholar]
- 18.del Campo J, et al. Taming the smallest predators of the oceans. ISME J. 2013;7:351–358. doi: 10.1038/ismej.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon HS, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 20.Massana R, et al. Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J. 2013;8:854–866. doi: 10.1038/ismej.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.del Campo J, Massana R. Emerging diversity within Chrysophytes, Choanoflagellates and Bicosoecids based on molecular surveys. Protist. 2011;162:435–448. doi: 10.1016/j.protis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Guillou L, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ. Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 23.Stepanauskas R. Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Gachon CMM, et al. The Culture Collection of Algae and Protozoa (CCAP): a biological resource for protistan genomics. Gene. 2007;406:51–57. doi: 10.1016/j.gene.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Vaulot D, et al. The Roscoff Culture Collection (RCC): a collection dedicated to marine picoplankton. Nova Hedwigia. 2004;79:49–70. [Google Scholar]
- 26.Andersen R, et al. Provasoli-Guillard National Center for culture of marine phytoplankton 1997 list of strains. J. Phycol. 1997;33(s6):1–75. [Google Scholar]
- 27.Friedl T, Lorenz M. The Culture Collection of Algae at Göttingen University (SAG): a biological resource for biotechnological and biodiversity research. Procedia Environ. Sci. 2012;15:110–117. [Google Scholar]
- 28.Adl SM, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fast NM, et al. Re-examining alveolate evolution using multiple protein molecular phylogenies. J. Eukaryot. Microbiol. 2002;49:30–37. doi: 10.1111/j.1550-7408.2002.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 30.Smirnov A, et al. Molecular phylogeny and classification of the lobose amoebae. Protist. 2005;156:129–142. doi: 10.1016/j.protis.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. CRC Crit. Rev. Plant Sci. 2012;31:1–46. [Google Scholar]
- 32.Simpson AGB, Patterson DJ. The ultrastructure of Carpediemonas membranifera (Eukaryota) with reference to the ‘Excavate hypothesis’. Eur. J. Protistol. 1999;35:353–370. [Google Scholar]
- 33.Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic ‘supergroups’. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single copy protein domains. Mol. Biol. Evol. 2011;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burki F, Keeling PJ. Rhizaria. Curr. Biol. 2014;24:103–107. doi: 10.1016/j.cub.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Burki F, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riisberg I, et al. Seven gene phylogeny of heterokonts. Protist. 2009;160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Edman JC, et al. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 39.Thines M, Kamoun S. Oomycete-plant coevolution: recent advances and future prospects. Curr. Opin. Plant. Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 41.Sebé-Pedrós A, et al. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sebé-Pedrós A, et al. Evolution and classification of myosins, a paneukaryotic whole genome approach. Genome Biol. Evol. 2014;6:290–235. doi: 10.1093/gbe/evu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Not F, et al. New insights into the diversity of marine picoeukaryotes. PLoS ONE. 2009;4:7. doi: 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.