Abstract
The eukaryotic supergroup Opisthokonta includes animals (Metazoa), fungi, and choanoflagellates, as well as the lesser known unicellular lineages Nucleariidae, Fonticula alba, Ichthyosporea, Filasterea and Corallochytrium limacisporum. Whereas the evolutionary positions of the well-known opisthokonts are mostly resolved, the phylogenetic relationships among the more obscure lineages are not. Within the Unikonta (Opisthokonta and Amoebozoa), it has not been determined whether the Apusozoa (apusomonads and ancyromonads) or the Amoebozoa form the sister group to opisthokonts, nor to which side of the hypothesized unikont/bikont divide the Apusozoa belong. Aiming at elucidating the evolutionary tree of the unikonts, we have assembled a dataset with a large sampling of both organisms and genes, including representatives from all known opisthokont lineages. In addition, we include new molecular data from an additional ichthyosporean (Creolimax fragrantissima) and choanoflagellate (Codosiga botrytis). Our analyses show the Apusozoa as a paraphyletic assemblage within the unikonts, with the Apusomonadida forming a sister group to the opisthokonts. Within the Holozoa, the Ichthyosporea diverge first, followed by C. limacisporum, the Filasterea, the Choanoflagellata, and the Metazoa. With our data enriched tree, it is possible to pinpoint the origin and evolution of morphological characters. As an example, we discuss the evolution of the unikont kinetid.
Keywords: cilia, Corallochytrea, eukaryotic phylogeny, Filasterea, Opisthokonts, Unikonta
Introduction
Our understanding of organismal evolution has improved significantly in recent decades, thanks largely to the contributions of improved molecular techniques and new microscopy data. Molecular phylogenies have consistently improved the tree of eukaryotes, now divided into five or six supergroups (Simpson and Roger 2004; Adl et al. 2005; Keeling et al. 2005; Roger and Simpson 2009). However, the root of the eukaryotes and the relationships among these eukaryotic supergroups remain uncertain. One of the most widespread hypotheses roots the eukaryote tree between the Unikonta, unicellular organisms whose ancestral mode of locomotion appears to have been based on a single cilium and basal body, and the Bikonta (Cavalier-Smith 2002). Other authors have proposed alternative hypotheses, placing the root of the eukaryotes in the Excavata (Cavalier-Smith 2010) or the Plantae (Rogozin et al. 2009); however, these hypotheses do not generally contradict a monophyletic union of the unikonts (except see Katz et al. 2012).
The unikonts, comprised of the Amoebozoa and Opisthokonta, have a striking diversity of forms. For example, several types of multicellularity have independently emerged, including the distinctive and well known metazoan and fungal body plans, the colonial stages of choanoflagellates and ichthyosporeans, and the aggregative fruiting bodies of Fonticula alba and dictyostelid slime molds (Paps and Ruiz-Trillo 2010 and references within). A sister group relationship between the Amoebozoa and the Opisthokonta has been supported by several molecular phylogenies (Burki et al. 2007; Rodriguez-Ezpeleta et al. 2007; Ruiz-Trillo et al. 2006; Ruiz-Trillo et al. 2008) and molecular synapomorphies (Richards and Cavalier-Smith 2005; Rogozin et al. 2009; Stechmann and Cavalier-Smith 2002). However, the Apusozoa, which consists of apusomonads and ancyromonads, has a contentious relationship with the unikonts. In some studies the Apusomonadida appear to belong within the unikonts and to be related to the opisthokonts (Cavalier-Smith and Chao 1995, Kim et al. 2006, Katz et al. 2011, Torruella et al. 2012). Other data, however, suggests a closer relationship to the bikonts due to their bi-flagellated form and the presence of a bikont-specific molecular gene fusion (Stechmann and Cavalier-Smith 2002; Stechmann and Cavalier-Smith 2003).
The monophyletic grouping of Opisthokonta is well supported by both molecular trees (Baldauf et al. 2000; Lang et al. 2002; Medina et al. 2003; Ruiz-Trillo et al. 2004; Ruiz-Trillo et al. 2008; Steenkamp and Baldauf 2004; Steenkamp et al. 2006; Torruella et al. 2012) and molecular synapomorphies, such as a 12 amino acid insertion in the elongation 1 alpha (EF1-alpha) gene (Baldauf and Palmer 1993; Steenkamp and Baldauf 2004) and a haloarchaeal-type tyrosyl tRNA synthetase (Huang et al. 2005, but see Shadwick and Ruiz-Trillo 2012). These molecular analyses also tend to divide the opisthokonts into two clades: the Holozoa (Lang et al. 2002), which includes the Metazoa and their unicellular relatives, and the Holomycota (Liu et al. 2009, also named Nucletmycea (Brown et al. 2009), which contains the fungi, nucleariids, and F. alba. Within the Holozoa, the sister-group relationship of Metazoa and choanoflagellates is also well supported (Carr et al. 2008; Lang et al. 2002; Medina et al. 2001; Ruiz-Trillo et al. 2006; Ruiz-Trillo et al. 2008; Torruella et al. 2012).
Within the unikonts the relationships between several lineages and placement of certain enigmatic taxa remain undetermined. For example, within the Holozoa, the position of C. limacisporum, relative to the Filasterea (Ministeria vibrans + Capsaspora owczarzaki) and Ichthyosporea is still contentious (reviewed by Paps and Ruiz-Trillo 2010), as are the specific relationships between the Holomycota lineages F. alba, Nucleariidae and Fungi. The affiliation of Breviata anathema (Walker et al. 2006), previously known as Mastigamoeba invertens (Minge et al. 2008), with the Amoebozoa is also uncertain. Some studies with high numbers of genes (Minge et al. 2008; Ruiz-Trillo et al. 2008; Shalchian-Tabrizi et al. 2008; Torruella et al. 2012) and others with fewer genes but broader taxonomic sampling (Brown et al. 2009; Ruiz-Trillo et al. 2004; Ruiz-Trillo et al. 2006; Steenkamp et al. 2006) have been able to hint at the positions of some of these organisms. However, no single molecular study so far has included multiple representatives from each of the known opisthokont lineages.
In this study we have built a dataset that balances both the sampling of taxa and markers with the aim of solving the position of the lesser-known taxa and clades in the unikont tree. Our alignment contains representatives of all the main opisthokont clades, several representatives of the Amoebozoa and the Apusozoa, and a robust selection of bikonts. We have collected data for eight molecular markers, the two ribosomal subunit rDNA genes (18S and 28S) and six protein coding genes: actin, elongation factor 1 alpha (EF1 alpha), alpha and beta tubulins, and the heat shock proteins 70 (hsp 70) and 90 (hsp 90). We have also produced new sequences by a PCR-based survey and by mining genome projects and ESTs (expressed sequence tag) collections. Specifically, we have amplified and sequenced five molecular markers from the ichthyosporean C. fragrantissima (Marshall et al. 2008) and the freshwater choanoflagellate C. botrytis. The final alignment contains 73 taxa and was analyzed using probabilistic methods.
Results
Datasets
The main molecular dataset contains 492 different sequences, and had 84% occupancy (i.e. 84% of all possible sequences for all taxa were sampled) and 30% missing data. Taxa used, their percentage of missing data, and their accession numbers are listed in Supplementary Table 1. There are 12 taxa whose missing data percentage is above 50%, and they are evenly spread through the groups sampled. The data consists of nucleotides from the two ribosomal RNA genes and amino acids from the 6 protein coding genes, totalling 6,110 characters. To test the effect of long branched Amoebozoa on the position of the Apusozoa, two additional datasets were generated, one excluding the longest branched amoebozoans and another with no amoebozoan representatives.
Phylogenetic analyses
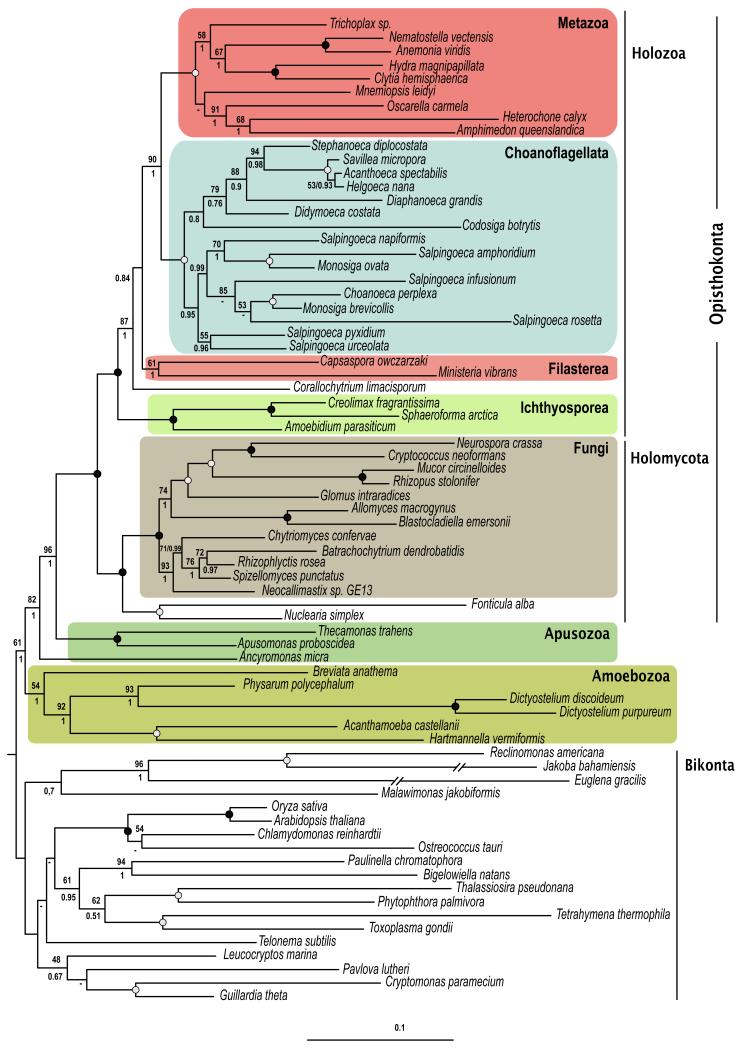
Figure 1 shows the combined results of the maximum likelihood (ML) and Bayesian inference (BI) analyses of our dataset. Both trees show almost the same branching pattern and display high support for several nodes. Notable exceptions include the weak association of B. anathema with Amoebozoa, a lack of support for a relationship between the Choanoflagellata and Filasterea, and poor resolution of some of the internal branches within the choanoflagellates. The datasets which either exclude all amoebozoans, including B. anathema (Figure S1), or exclude the fastest evolving Amoebozoa (Figure S2), show topologies mostly congruent with the main dataset, except that B. anathema groups with the bikonts in the analysis without the fastest evolving Amoebozoa (Figure S2).
Figure 1. ML phylogenetic tree of the Unikonta from eight genes.
Phylogenetic tree of the Unikonta inferred from the 8 markers by Maximum Likelihood . A black dot indicates a clade with Bayesian Posterior Probabilities (PP values of 1.0 and Bootstrap Support (BS of 100%. A grey dot indicates a clade with PP values of 1.0 and BS of 97-99%. Values above and below the branches correspond to BS values (if greater than 50%), and PP values, respectively. Dashes specify non recovered nodes. The scale bar indicates the number of changes per site. For accession numbers corresponding to the markers for each terminal see Supplementary Table 1.
The tree shows the division of Unikonta, including Apusozoa, from the Bikonta,(Figure 1), albeit with weak support. However, when amoebozoans are completely removed from the analysis (Figure S1), the support for the unikont-bikont partition increases to a bootstrap support (BS) of 94%. This may be caused by the unstable phylogenetic position of the B. anathema, which has 57% missing data, and/or the long branches present in most amoebozoan representatives. B. anathema is positioned as the sister group to all other amoebozoans, but with low statistical support (BS=54%). The monophyletic relationship and internal nodes of the other conventionally accepted amoebozoans are well supported.
The Apusozoa appear as a paraphyletic assemblage laddered between the amoebozoans and the opisthokonts (Figure 1). The apusozoan Ancyromonas micra (Order Ancyromonadida, formerly known as Planomonas micra belonging to Order Planomonadida, see Heiss et al. 2010) diverges first while a clade consisting of Apusomonas proboscidea and Thecamonas trahens (Order Apusomonadida, T. trahens was formerly known as Amastigomonas sp. ATCC50062, see Cavalier-Smith and Chao 2010) is shown as sister group to the opisthokonts. Both the relationship between the two apusozoan lineages and the opisthokonts and the split between the ancyromonads and apusomonads are further reinforced when the amoebozoans are removed from the analyses (Figure S1 ).
The Opisthokonta are recovered with maximum support with all the alignments and algorithms (Figures 1, S1 and S2). The subdivision into the Holozoa and the Holomycota is also well supported. Within the Holomycota, two clades with high statistical support are found, one leading to F. alba and Nuclearia simplex and the other to Fungi. Within the Holozoa, the trees show the Ichthyosporea (Amoebidium parasiticum, Sphaeroforma arctica and C. fragrantissima) as their first branch, with C. fragrantissima as a sister group to S. arctica (with maximum support by all analyses, Figures 1, S1 and S2). The node that positions the ichthyosporeans at the base of the Holozoa and groups C. limacisporum with the remaining Holozoa holds moderately high support (BS=87%), which increases in the tree without Amoebozoa (BS=98%, see Figure S1). While it seems clear that Corallochytrea, Filasterea and Choanoflagellates + Metazoa form a clade, the exact branching order between them is not resolved. Also, while the monophyly of Filasterea (M. vibrans and C. owczarzaki, Shalchian-Tabrizi et al. 2008) is recovered, the support values are not high (BS=61%). The Metazoa and the Choanoflagellata are strongly recovered as sister groups (BS=90%), and the internal phylogeny of the choanoflagellates recovers two clades present in previous studies (Nitsche et al. 2011): the Acanthoecidae and the Salpingoecidae (Figure 1). The position of the freshwater choanoflagellate C. botrytis grouped with the Acanthoecidae but only with moderate Bayesian support (Figure 1).
Topological constraint tests
Different phylogenetic hypotheses were statistically tested (Table 1). Most of the constrained topologies were based upon results from previous publications. Three hypothetical positions for the Apusozoa were tested: 1) the ‘Apusozoa as monophyletic’ hypothesis (Cavalier-Smith and Chao 2003), 2) the ‘Apusozoa as members of the Bikonta’ (Stechmann and Cavalier-Smith 2003), and 3) the ‘Apusozoa as a sister group to the Unikonta’ (where the Bikonta are also constrained as a monophyletic lineage). Within the opisthokonts several alternative branching orders were tested including: positioning C. limacisporum as a sister group to the Choanoflagellata (Cavalier-Smith and Chao 2003; Jostensen et al. 2002; Mendoza et al. 2002; Ruiz-Trillo et al. 2006), a C. limacisporum sister group relationship to the Ichthyosporea (Carr et al. 2008; Ruiz-Trillo et al. 2004; Ruiz-Trillo et al. 2006; Steenkamp et al. 2006), and an ichthyosporean sister group relationship to the Filasterea (similar to Ruiz-Trillo et al. (2008) where C. owczarzaki represents the Filasterea). The Approximately Unbiased test (AU test) results (Table 1) show that all these alternative hypotheses were statistically rejected (values under 0.05), except for the sister-group relationship of Filasterea and Ichthyosporea.
Table 1.
Comparison of topologies using the approximately unbiased test
| Topologies | AU Test (p-values) |
|---|---|
| Test 1 | |
| Best tree | 0.87 |
| Corallochytrea + Choanoflagellata | 0.0054 |
| Corallochytrea + Ichthyosporea | 0.013 |
| Ichthyosporea + Filasterea | 0.076 |
| Test 2 | |
| Best tree | 0.99 |
| Apusozoa monophyletic | 0.0056 |
| Apusozoa with Bikonta | 2.7e-04 |
| Apusozoa as a sister group to Unikonta | 4.0e-04 |
Discussion
Unikonta phylogeny
Our results recover the Amoebozoa, including B. anathema, as one of two major unikont clades (Figure 1), although this arrangement is not robustly supported. This result has also been seen in previous ribosomal (18S) and phylogenomic analyses (Nikolaev et al. 2006; Minge et al. 2008). Recent phylogenomics analyses have suggested that Breviatea (Cavalier-Smith et al. 2004) is the sister group to all other amoebozoans (Minge et al. 2008), but other analyses have shown B. anathema to be related to the Apusozoa (Walker et al. 2006; Katz et al. 2011). Our data can not confidently define whether B. anathema branches within or outside the Amoebozoa.
Our analyses show the Apusozoa as a paraphyletic grouping, with the Ancyromonadida (Heiss et al. 2010) splitting first, followed by the well supported Apusomonadida (Karpov and Mylnikov 1989) and Opisthokonta. Although this result was reinforced following removal of the amoebozoan taxa, it is based on a single representative of the Ancyromonadida. A recent article by Cavalier-Smith and Chao (2010) based on the 18S ribosomal gene also suggests, although with low support, that the apusozoans are paraphyletic and that they are more closely related to opisthokonts than to amoebozoans. In addition, Torruella et al. (2012) recovered a well supported sister group relationship with the opisthokonts and a single apusomonad representative (T. trahens), using phylogenomic analysis. Finally, our topological restraint test rejected all alternative hypotheses enforcing either a monophyletic union of the Apusozoa or their position elsewhere in the eukaryote tree (Table 1).
The Opisthokonta
The Opisthokonta and their division into two major subgroups, the Holomycota and the Holozoa, are strongly supported by all our analyses (Figures 1, S1 and S2). While those two subgroups have been recovered in previous trees (Lang et al. 2002; Medina et al. 2003; Ruiz-Trillo et al. 2004; Steenkamp and Baldauf 2004; Ruiz-Trillo et al. 2008;), this is the first multigenic analyses to include such high numbers of choanoflagellates and ichthyosporeans, as well as generous taxon sampling from close outgroup taxa. Within the Holomycota, N. simplex and F. alba form a robust clade as a sister group to the Fungi, in agreement with the only other study with comparable taxon sampling (Brown et al. 2009) (Figure 1).
Within the Holozoa, our results support the well-known relationship between the Metazoa and the Choanoflagellata (Carr et al. 2008; Lang et al. 2002; Medina et al. 2001; Ruiz-Trillo et al. 2006; Ruiz-Trillo et al. 2008; Torruella et al. 2012). Most of the internal phylogeny of Choanoflagellata is weakly supported, likely a consequence of the high percentage of missing data in most choanoflagellate taxa. Our trees support a sister-group relationship between the Ichthyosporea and the rest of holozoans rather than as a sister group to the Filasterea alone as shown by Ruiz-Trillo et al. (2008). This result is more consistent with the “Filozoa hypothesis” (Shalchian-Tabrizi et al. 2008) as seen in other molecular analyses (Ruiz-Trillo et al. 2008; Shalchian-Tabrizi et al. 2008; Torruella et al. 2012), but the relationship between Ichthyosporea and Filasterea can not be rejected by our AU test (p-value 0,076).
Our analysis differs in that it also includes C. limacisporum. C. limacisporum has previously been suggested to be related to the choanoflagellates (Cavalier-Smith and Allsopp 1996), to Fungi (Sumathi et al. 2006) or to the Ichthyosporea (Steenkamp et al. 2006). Instead our results suggest that C. limacisporum is a sister lineage to the Filazoa and all previous hypotheses were rejected based on the AU test of our dataset. Nonetheless, the specific position of C. limacisporum remains unresolved, and more data and wider taxon sampling are almost certainly needed to recover this specific phylogenetic position. Unfortunately current taxon sampling from Corallochytrium, Capsaspora, and Ministeria, is limited by the number of representatives known to science.
The evolution of basal body arrangement and the unikont/bikont condition
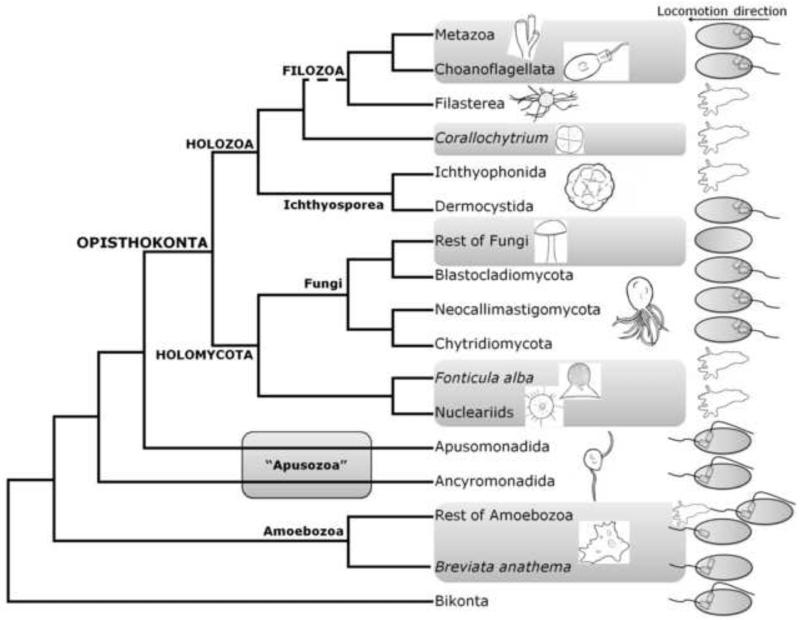
Although our tree does not robustly infer the phylogenetic position of all taxa analyzed, it does both resolve and confirm many relationships, thereby forming a reasonable starting point to postulate evolutionary scenarios. As an example, Figure 2 shows a summary of our main results, accompanied by a sketch of the locomotion complex (kinetid) for each group. This scheme can be used to outline a hypothetical proposal for the evolution of the flagellar apparatus in unikonts. Nonetheless, we recognize that this proposal is limited by both the number species known to date, as well as the incomplete morphological data available. The kinetid is formed by one or two flagella and one or more basal bodies. Flagella are always attached to a basal body, but not all the basal bodies within a kinetid have an attached flagella. Groups that lack a kinetid tend to move by amoeboid movements (see Figure 2). The structure of the kinetid in the last common ancestor of all Amoebozoa is important for understanding the origins of the eukaryotes, i.e. the validity of unikont/bikont rooting. Although most amoebozoans do not have flagella, some display one or more flagella and varying numbers of basal bodies. For example, one basal body is found in some Archamoebae and in the conosean Multicilia marina, two basal bodies are seen in some myxogastrids slime molds, and a mixture of both states are found in protostelids (Minge et al. 2008). The kinetid configuration consisting of one flagellum and one basal body as displayed in some amoebozoans is similar to the ancestral unikont-like eukaryote proposed by Cavalier-Smith (2002); thus an ancestral Amoebozoa with this arrangement would support the latter hypothesis. However, those flagellated amoebozoans having a single basal body show a more recently derived position within the amoebozoans, making it difficult to clarify the ancestral state of the group. Furthermore, B. anathema, which positions here as the sister group to all other amoebozoans, has a single flagellum but has at least two basal bodies, a state also present in the flagellated Opisthokonta (Roger and Simpson 2009). In contrast to opisthokonts, B. anathema’s kinetid is found in the anterior part of the cell while that of opisthokonts’ is in the posterior part. This, together with B. anathema’s position as the earliest known branch of Amoebozoa (Figure 1, Minge et al. 2008), points to the presence of at least two basal bodies in the unikont’s last common ancestor, as opposed to an ancestor with a single basal body, and further suggests that the kinetid was located in the anterior part of the cell. This hypothesis is further reinforced by the apparent paraphyly of the Apusozoa, where each branch is represented by taxa having two flagella and two basal bodies in the anterior part of the cell (Roger and Simpson 2009). Therefore the presence of an anterior kinetid with at least two basal bodies in some bikonts, B. anathema, ancyromonads and apusomonads, suggests that the last common ancestor of eukaryotes may also have had an anterior kinetid with at least two basal bodies.
Figure 2. Schematic tree of the unikonts with an outline of the kinetid structure.
Inferring the number of flagella per kinetid in the last common eukaryotic ancestor is more speculative and may involve several independent gains and/or losses. Assuming that the ancestor had two flagella (as in bikonts and the two apusozoan branches), then one flagellum was independently lost in Breviatea and Opisthokonta. Alternatively, if the eukaryotic ancestor had one flagellum, then a second one was independently gained within the Amoebozoa (i.e. in Physarium) and again, either in the last common ancestor of ancyromonads, apusomonads and opisthokonts (then lost again in opishtokonts) or in both the ancyromonads and apusomonads.
Within the Opisthokonta, the archetypical posteriorly flagellated morphology is represented by the choanoflagellates. Uni-flagellated life stages are present as zoospores in early-branching fungi (Tanabe et al. 2005) and the ichthyosporean order Dermocystida (Mendoza et al. 2002), and also as metazoan sperm cells. While it is clear that the kinetid was lost in the ichthyosporean order Ichthyophonida and in the derived fungi (Tanabe et al. 2005), the evolution of the flagellar apparatus in other opisthokonts remains less clear due to the limited number of isolated taxa and morphological studies.
If the flagellar apparatus is homologous between opisthokonts and apusozoans, then the possible paraphyly of the apusozoans would indicate an apusozoan-like ancestor for opisthokonts. This allows for a provisional description of kinetid evolution in the opisthokont lineage. Accordingly, the anterior kinetid seen in apusozoans would have moved to the posterior part of the cell and one of the two flagella would have been lost, thereby leaving the opisthokont ancestor with a single posterior flagellum attached to one basal body and one non-flagellated basal body. This means that this second basal flagellum would have been lost twice within the unikonts: once in the anterior kinetid of B. anathema and another time in the opisthokont ancestor. Finally, the kinetid would have been lost on at least five occasions within the opisthokonts: 1) in the common ancestor of nucleariids and F. alba as proposed by Brown et al. (2009), 2) in the derived fungi, likely many times, consistent with Hibbet et al. (2007), and three more times within the Holozoa in 3) Ichthyophonida, 4) C. limacisporum and 5) Filasterea. In the case of filastereans, it is noteworthy that according to Cavalier-Smith (Cavalier-Smith and Chao 2003) “Ministeria vibrans has a vibratile stalk which has some ultrastructural similarity to a cilium”, but there is no evidence of flagellum/cilium in Ministeria marisola or C. owczarzaki. In any case, further data on the ontogeny, structure and genetics of the different eukaryotic clades is needed to completely understand flagellum evolution.
Conclusions
Our multigene analysis of the Opisthokonta confirms some previous phylogenetic relationships, such as the monophyly of the opisthokonts, the division of opisthokonts into Holozoa and Holomycota, and the monophyly of nucleariids + F. alba and their sister group relationship to Fungi. Our data also provide strong support to some contentious hypotheses, such as apusozoan paraphyly and their sister group relationship to opisthokonts, and moderate support to the position of Filasterea as sister group to Choanoflagellata and Metazoa rather than to Ichthyosporea. Finally, our results suggest that C. limacisporum is related to the filozoan clades, although its specific phylogenetic position within the Holozoa remains unclear. Our tree provides a framework on which to discuss character evolution as more morphological and biochemical data on lesser known taxa and lineages becomes available.
Methods
Organisms and molecular methods
The culture of the ichthyosporean C. fragrantissima strain CH2 was grown as specified in Marshall et al. (2008). The culture of the choanoflagellate C. botrytis was a kind donation from Prof. Hartmut Arndt (University of Cologne) to Hiroshi Suga and was grown in WC medium (Guillard and Lorenzen 1972). The RNA extractions were performed from pelleted live cultures using TRIzol (Amersham Pharmacia Biotech) and cDNA was obtained by reverse transcription with M-MLV reverse transcriptase (Promega). Eight markers were amplified and sequenced: the two ribosomal subunit rDNA genes (18S and 28S) and six protein coding genes: actin, elongation factor 1 alpha (EF1 alpha), alpha and beta tubulins, and the heat shock proteins 70 (hsp 70) and 90 (hsp 90). The 8 gene fragments were amplified by PCR using universal degenerate primers (Supplementary Table 2). The PCR was performed with 25 μl final volume, using 1 unit of Taq polymerase and 0.5 μl betaine (Sigma) as PCR enhancer. PCR products were purified using Microcon PCR columns (Millipore) and directly sequenced from both strands (Big Dye Terminator V.2.0, Applied Biosystems). Sequence products were ethanol precipitated and run on an ABI Prism 3700 (Applied Biosystems) automated sequencer. The sequence data were assembled with SeqEd V.1.0.3 (Applied Biosystems). New sequences have been deposited in GenBank with accession numbers HQ896013-22.
Alignments and Phylogenetic Analyses
Gene sequences were downloaded from GenBank or from genome and EST projects hosted in different public institutes (Broad Institute, Joint Genome Institute, etc). See Supplementary Table 1 for sequences origins and accession numbers. Each gene was aligned independently using MAFFT (Katoh et al. 2005), and the resulting alignments were checked by eye with Bioedit v. 7.0.9.0 (Hall 1999). Regions of ambiguous alignment were removed using the online version of Gblocks (Castresana 2000) with the “less stringent selection” options selected. The final alignments had 1390 positions for 18S, 2206 for 28S, 371 for actin, 404 for EF1-alpha, 508 for hsp70, 534 for, hsp90, 392 for tubulin alpha, and 398 for tubulin beta. For each alignment, gene orthology was assessed with single-gene phylogenies. The final curated alignments were concatenated using Bioedit, producing a 6,110 positions matrix for 73 taxa. Alignments can be downloaded from the webpage http:www.multicellgenome.com and have been deposited at Dryad Repository: http://dx.doi.org/10.5061/dryad.v3p3j.
Bayesian inference trees were inferred with a parallelized version of MrBayes software v.3.1.2 (Ronquist and Huelsenbeck 2003), using a partitioned dataset (one partition for each gene, unlinking parameter estimation for each partition) and running 3,000,000 generations in 2 independent sets of 4 chains (with a sample frequency of 1 in 1000). MrBayes was run using the model GTR + Γ + I (4 gamma categories + 1 invariable) for the nucleotide markers (ribosomal genes) and WAG + Γ + I (4 gamma categories + 1 invariable) for the protein-coding genes; all the 8 partitions had an independently applied co-varion correction. Before obtaining consensus tree and posterior probabilities, all trees sampled before the likelihood values reached a plateau were removed, resulting in a burnin of a 1,000,000 generations (all the PRSF values approaching 1 to the second decimal point). Maximum likelihood trees were inferred with RAxML v.7.2.6 (Stamatakis 2006), using a partitioned dataset (8 partitions, one for each gene), using the model GTR + Γ + I (4 gamma categories + 1 invariable) for the nucleotide partitions (ribosomal genes) and LG + Γ + I (4 gamma categories + 1 invariable) for the amino acid ones (protein-coding genes). A random topology was used as starting tree and 1,000 bootstrap replicates were obtained (options -f i -b 1999 -#1000). Four independent analyses were run to check that the resulting topology was not the result of a single run being trapped in a local maxima. In order to evaluate the competing topologies, we performed two approximately unbiased tests (Shimodaira and Hasegawa 2001) on two topology sets. First (Test 1), the Holozoa phylogeny was analysed. All possible topologies were explored by combining seven subtrees (Metazoa, Choanoflagellata, Filasterea, Corallochytrea, Ichthyosporea, Fungi, and Apusozoa+Amoebozoa+Bikonta) from the ML tree, and explored all possible combinations of the sub-trees (745 topologies in total). Second (Test 2), the Apusozoa phylogeny was analysed. We tested all the possible positions, except inside the Holozoa, of the T. trahens + A. proboscidea lineage and the A. micra lineage on the ML toplogy, fixing the other branching patterns (2499 topologies in total). CONSEL V0.1i (Shimodaira and Hasegawa 2001) was used to perform the AU tests (Shimodaira 2002).
Supplementary Material
File S2. ML Phylogenetic tree inferred without the fastest-evolving Amoebozoa.
Phylogenetic tree inferred without the fastest-evolving Amoebozoa taxa (D. discoideum and D. purpureum ) by Maximum Likelihood. Only Bootstrap Support (BS values above 50% are indicated in the nodes.
File S4. List of primers used.
List of primers and the papers from where they were acquired. Table supplied as an Excel file.
File S1. ML Phylogenetic tree without Amoebozoa
Phylogenetic tree inferred without the Amoebozoa taxa by Maximum Likelihood. Only Bootstrap Support (BS values above 50% are indicated in the nodes. Note that the statistical support for the position of Apusozoa has slightly increased compared to Figure 1.
File S3. Names of the taxa and genes, together with their accession numbers.
Names of the taxa and genes, together with their accession numbers, considered in this study. Bold numbers indicate new sequences obtained by PCR in this study. Table supplied as an Excel file.
Acknowledgements
We are grateful to Arnau Sebé, Alex de Mendoza, Romain Derelle and Guifré Torruella for all the lively discussions and stimulating insights on the subject. We thank Martin Carr’s (Faculty of Biological Sciences, University of Leeds) feedback on the internal choanoflagellate phylogeny, Harmut Arndt (Zoological Institute, University of Cologne) for sharing Codosiga botrytis culture, B. Franz Lang (Département de Biochimie, Université de Montréal) for access to Thecamonas trahens genome sequences, as well as Alastair G. Simpson (Department of Biology, Dalhousie University) and Thomas Cavalier-Smith (Department of Zoology, University of Oxford) for their helpful feedback on flagellar apparatus across different groups. The authors thank Prof. Peter Holland (Department of Zoology, University of Oxford) for his support and the 2 anonymous referees for the great feedback provided. We would like to thank Maria José Barberà for the graphical support and Sara Rojas for the animal drawings included in Figure 2. The authors thankfully acknowledge the computer resources, technical expertise and assistance provided by the Barcelona Supercomputing Center- Centro Nacional de Supercomputación, specially to David Vicente (Barcelona Supercomputing Centre) and Luis Cabellos (Instituto de Física de Cantabria). H.S. is supported by the Marie Curie Intra-European Fellowship within the 7th European Community Framework Programme. This work was supported by an ICREA contract, an European Research Council Starting Grant (ERC-2007-StG-206883), and two grants (BFU2008-02839/BMC and BFU2011-23434) from Ministerio de Ciencia e Innovación (MICINN) to IR-T.
References
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 2009;26:2699–2709. doi: 10.1093/molbev/msp185. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci USA. 2008;105:16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The origin of Fungi and Pseudofungi. In: ADM Rayner, CM Brasier, D Moore., editors. Evolutionary Biology of the Fungi. Cambridge University Press; Cambridge, United kingdom: 1987. pp. 339–353. [Google Scholar]
- Cavalier-Smith T, Chao E. The Opalozoan Apusomonas is related to the common ancestor of Animals, Fungi, and Choanoflagellates. ProcR Soc B. 1995;261:1–6. [Google Scholar]
- Cavalier-Smith T, Allsopp MTEP. Corallochytrium, an enigmatic non-flagellate protozoan related to choanoflagellates. Eur J Protistol. 1996;32:306–310. [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny of Choanozoa, Apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56:540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao E, Oates B. Molecular phylogeny of Amoebozoa and the evolutionary significance of the unikont Phalansterium. Eur J Protistol. 2004;40:21–48. [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and Evolution of Apusomonadida (Protozoa: Apusozoa): new genera and species. Protist. 2010;161:549–576. doi: 10.1016/j.protis.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–345. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL, Lorenzen CJ. Yellow-green algae with chlorophyllide C1, 2. J Phycol. 1972;8:10–14. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Heiss AA, Walker G, Simpson AG. Clarifying the taxonomic identity of a phylogenetically important group of eukaryotes: Planomonas is a junior synonym of Ancyromonas. J Eukaryot Microbiol. 2010;57:285–293. doi: 10.1111/j.1550-7408.2010.00477.x. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Koljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schussler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Huang J, Xu Y, Gogarten JP. The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Mol Biol Evol. 2005;22:2142–2146. doi: 10.1093/molbev/msi221. [DOI] [PubMed] [Google Scholar]
- Jostensen J-P, Sperstad S, Johansen S, Landfald B. Molecular-phylogenetic, structural and biochemical features of a cold-adapted, marine ichthyosporean near the animal-fungal divergence, described from in vitro cultures. Eur J Protistol. 2002;38:93–104. [Google Scholar]
- Karpov SA, Mylnikov AP. Biology and ultrastructure of colourless flagellates Apusomonadida ord. n. Zoologischkei Zhurnal. 1989;58:5–17. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Grant J, Parfrey LW, Gant A, O’Kelly CJ, Anderson OR, Molestina RE, Nerad T. Subulatomonas tetraspora nov. gen. nov. sp. is a member of a previously unrecognized major clade of eukaryotes. Protist. 2011;162:762–773. doi: 10.1016/j.protis.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Katz LA, Grant JR, Parfrey LW, Burleigh JG. Turning the crown upside down: gene tree parsimony roots the eukaryotic tree of life. Syst Biol. 2012 doi: 10.1093/sysbio/sys026. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kim E, Simpson AG, Graham LE. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol. 2006;23:2455–2466. doi: 10.1093/molbev/msl120. [DOI] [PubMed] [Google Scholar]
- Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- Liu Y, Steenkamp ET, Brinkmann H, Forget L, Philippe H, Lang BF. Phylogenomic analyses predict sistergroup relationship of nucleariids and fungi and paraphyly of zygomycetes with significant support. BMC Evol Biol. 2009;9:272. doi: 10.1186/1471-2148-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WL, Celio G, McLaughlin DJ, Berbee ML. Multiple isolations of a culturable, motile ichthyosporean (Mesomycetozoa, Opisthokonta), Creolimax fragrantissima n. gen., n. sp., from marine invertebrate digestive tracts. Protist. 2008;159:415–433. doi: 10.1016/j.protis.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Collins AG, Taylor JW, Valentine JW, Lipps JH, AmaralBZettler L, Sogin ML. Phylogeny of Opisthokonta and the evolution of multicellularity and complexity in Fungi and Metazoa. Int J Astrobiol. 2003;2:203–211. [Google Scholar]
- Mendoza L, Taylor JW, Ajello L. The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu Rev Microbiol. 2002;56:315–344. doi: 10.1146/annurev.micro.56.012302.160950. [DOI] [PubMed] [Google Scholar]
- Minge MA, Silberman JD, Orr RJ, Cavalier-Smith T, Shalchian-Tabrizi K, Burki F, Skjaeveland A, Jakobsen KS. Evolutionary position of breviate amoebae and the primary eukaryote divergence. Proc Biol Sci. 2008;276:597–604. doi: 10.1098/rspb.2008.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Berney C, Petrov NB, Mylnikov AP, Fahrni JF, Pawlowski J. Phylogenetic position of Multicilia marina and the evolution of Amoebozoa. Int J Syst Evol Microbiol. 2006;56:1449–1458. doi: 10.1099/ijs.0.63763-0. [DOI] [PubMed] [Google Scholar]
- Nitsche F, Carr M, Arndt H, Leadbeater BS. Higher level taxonomy and molecular phylogenetics of the choanoflagellatea. J Eukaryot Microbiol. 2011;58:452–462. doi: 10.1111/j.1550-7408.2011.00572.x. [DOI] [PubMed] [Google Scholar]
- Paps J, Ruiz-Trillo I. Encyclopedia of Life Sciences (ELS) John Wiley & Sons, Ltd; Chichester: 2010. Animals and Their Unicellular Ancestors. [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Brinkmann H, Burger G, Roger AJ, Gray MW, Philippe H, Lang BF. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Simpson AG. Evolution: revisiting the root of the eukaryote tree. Curr Biol. 2009;19:R165–7. doi: 10.1016/j.cub.2008.12.032. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Basu MK, Csuros M, Koonin EV. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol Evol. 2009;1:99–113. doi: 10.1093/gbe/evp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxford, England) 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Inagaki Y, Davis LA, Sperstad S, Landfald B, Roger AJ. Capsaspora owczarzaki is an independent opisthokont lineage. Curr Biol. 2004;14:R946–947. doi: 10.1016/j.cub.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Lane CE, Archibald JM, Roger AJ. Insights into the evolutionary origin and genome architecture of the unicellular opisthokonts Capsaspora owczarzaki and Sphaeroforma arctica. J Eukaryot Microbiol. 2006;53:1–6. doi: 10.1111/j.1550-7408.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- Shadwick JD, Ruiz-Trillo I. A genomic survey shows that the haloarchaeal type tyrosyl tRNA synthetase is not a synapomorphy of opisthokonts. Eur J Protistol. 2012;48:89–93. doi: 10.1016/j.ejop.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS ONE. 2008;3:e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics (Oxford, England) 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–6. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13:R665–6. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Baldauf SL. Origin and Evolution of animals, fungi and their unicellular allies (Opisthokonta) In: Hirt RP, Horner DS, editors. Organelles, genomes and eukaryote phylogeny: an evolutionary synthesis in the age of genomics. CRC Press; Boca Raton: 2004. pp. 109–129. [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Sumathi JC, Raghukumar S, Kasbekar DP, Raghukumar C. Molecular evidence of fungal signatures in the marine protist Corallochytrium limacisporum and its implications in the evolution of animals and fungi. Protist. 2006;157:363–376. doi: 10.1016/j.protis.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, Watanabe MM, Sugiyama J. Evolutionary relationships among basal fungi (Chytridiomycota and Zygomycota): Insights from molecular phylogenetics. J Gen Appl Microbiol. 2005;51:267–276. doi: 10.2323/jgam.51.267. [DOI] [PubMed] [Google Scholar]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G, Dacks JB, Martin Embley T. Ultrastructural description of Breviata anathema, n. gen., n. sp., the organism previously studied as “Mastigamoeba invertens”. J Eukaryot Microbiol. 2006;53:65–78. doi: 10.1111/j.1550-7408.2005.00087.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S2. ML Phylogenetic tree inferred without the fastest-evolving Amoebozoa.
Phylogenetic tree inferred without the fastest-evolving Amoebozoa taxa (D. discoideum and D. purpureum ) by Maximum Likelihood. Only Bootstrap Support (BS values above 50% are indicated in the nodes.
File S4. List of primers used.
List of primers and the papers from where they were acquired. Table supplied as an Excel file.
File S1. ML Phylogenetic tree without Amoebozoa
Phylogenetic tree inferred without the Amoebozoa taxa by Maximum Likelihood. Only Bootstrap Support (BS values above 50% are indicated in the nodes. Note that the statistical support for the position of Apusozoa has slightly increased compared to Figure 1.
File S3. Names of the taxa and genes, together with their accession numbers.
Names of the taxa and genes, together with their accession numbers, considered in this study. Bold numbers indicate new sequences obtained by PCR in this study. Table supplied as an Excel file.