Figure 1.
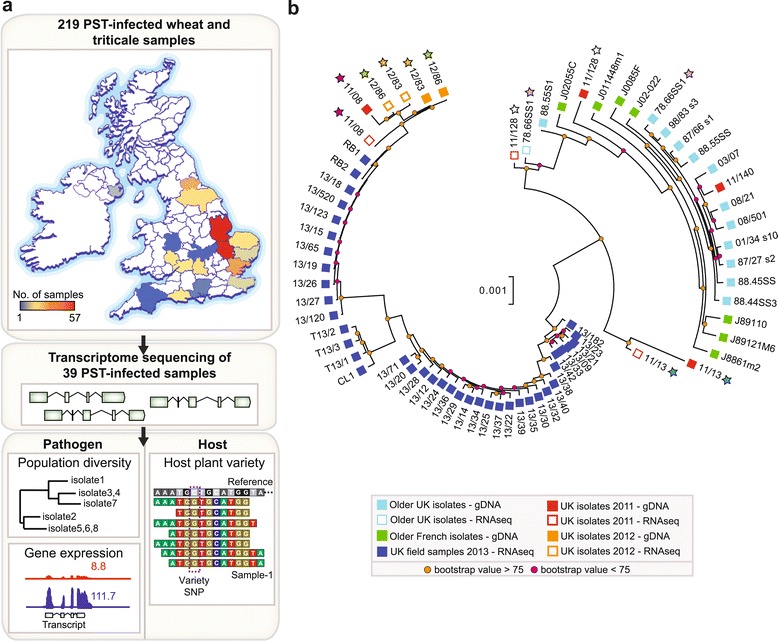
PST field isolates belong to a diverse emergent lineage. (a) A total of 219 samples of wheat and triticale infected with PST were collected from 17 different counties across the United Kingdom (UK) in the spring and summer of 2013. Transcriptome sequencing was carried out on 39 samples to generate transcript data from both the pathogen and host. For the pathogen, the data were used to assess the pathogen population diversity and differential gene expression. For the host, the data were used to confirm the host variety within a particular sample. SNP, single nucleotide polymorphism. (b) 2013 field isolates (dark blue squares) are distinct and highly diverse when compared with the older UK population (light blue squares). Phylogenetic analysis was undertaken using the third codon position of 5,610 PST-130 gene models (2,496,679 sites) with ≥80% breadth of coverage for all PST isolates using a maximum likelihood model. Stars indicate samples in which both the genome and transcriptome were sequenced from the same PST isolate.
