Figure 3.
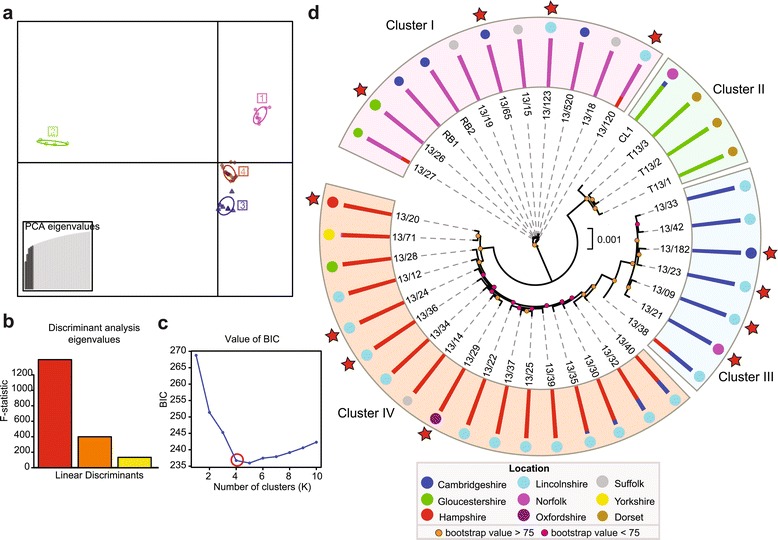
The 2013 PST field isolates are highly diverse and group genetically into four distinct population clusters. (a) Scatterplot using the first two principal components (Y-axis and X-axis, respectively) of the discriminant analysis of principal components (DAPC) analysis of 34,806 synonymous single nucleotide polymorphism (SNP) sites. Each symbol represents a single PST isolate, coloured according to assignment to one of four population clusters. All four population clusters are clearly separated by DAPC analysis. (b) The first three eigenvalue components from the DAPC analysis, supporting the maintenance of three discriminant functions in the DAPC analysis. (c) The optimal predicted number of population clusters K for the dataset is four. The Y-axis corresponds to the Bayesian information criterion (BIC), a goodness-of-fit measurement calculated for each K. The elbow in the BIC values (K = 4) indicates the optimal number of populations. (d) Phylogenetic analysis using a maximum likelihood model (2,513,246 sites) and Bayesian-based clustering of 34,806 synonymous SNP sites classified PST field isolates into four population clusters. All PST isolates sampled from triticale clustered within a single genetically distinct lineage, cluster II. Bar charts represent STRUCTURE analysis, with each bar representing estimated membership fractions for each individual. Stars highlight isolates purified for virulence profiling. Coloured circles represent UK counties in which samples were collected (Table S1 in Additional file 1).
