Figure 4.
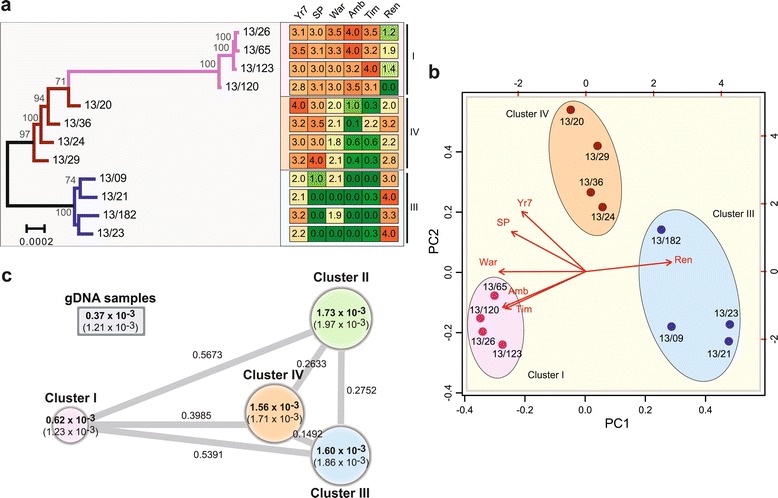
The 2013 PST field isolates identified on wheat show correlation between genetic and phenotypic profiles. (a) The population substructure illustrated by phylogenetic analysis of 12 PST isolates using a maximum likelihood model (6,479 genes; 2,792,462 sites) correlated with the virulence profiles of the isolates. Virulence profiles for each population cluster were largely conserved between members of the same population cluster, but differed from those of members of other population clusters. Four PST isolates from each of the three population clusters derived from PST-infected wheat samples were inoculated on a series of 39 different wheat varieties and the six discriminative results are shown: Yr7, AvocetS-Yr7 near isogenic line; Sp, Spaldings Prolific; War, Warrior; Amb, Ambition; Tim, Timber; Ren, Rendezvous. Disease severity was recorded 16 to 20 days post-inoculation: 0 (green) to 4 (red) scale with 0 to 2 resistant and 2 to 4 susceptible. Roman numerals represent population clusters. (b) Principal component analysis of the virulence profiles of the 12 PST isolates supports the separation of isolates into three distinct groups based on their phenotypes, which correlates with their genetic partitioning. (c) The degree of genetic diversity between members of a single population cluster for all 2013 PST field samples was much greater than that displayed by older UK isolates collected between 1978 and 2011. Circle size represents the degree of nucleotide diversity enclosed; standard deviation is given in parentheses. Branch lengths represent F ST values stated alongside.
