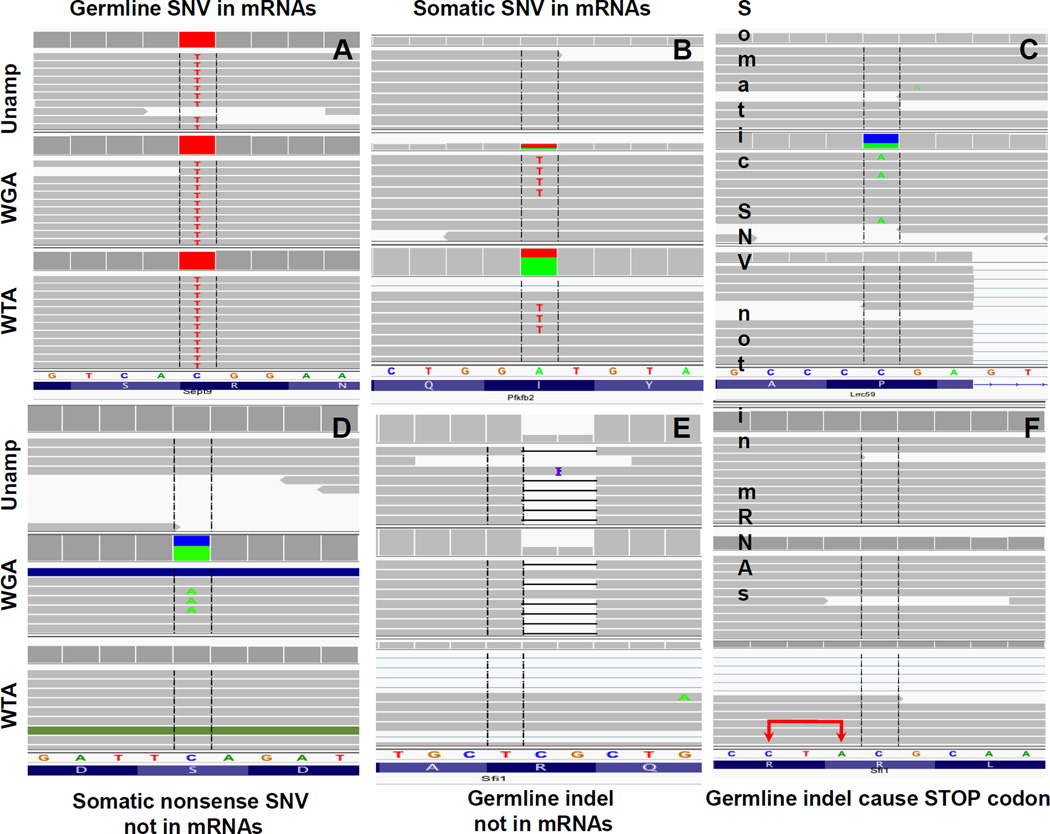
Figure 4. Integrative genomics viewer (IGV) images comparing specific mutations present in both exome and transcriptome from the same single cell.
The genetic background is defined by the unamplified control (Unamp). (A) The presence of a germline SNV in mRNAs. (B) The presence of a heterozygous somatic A:T transversion, a signature ENU-induced mutation, in mRNAs. (C) A heterozygous somatic SNV absent from mRNAs. (D) A somatic C to A transversion generating a nonsense mutation; no mutant transcript detected. (E) A heterogyzous bi-nucleotidyl CG-deletion absent from mRNAs. (F) The binucleotidyl deletion induces a premature TAG (CTA in the illustration; Sfi1 gene is transcribed in reversed order) stop codon (red arrows) in a downstream exon.