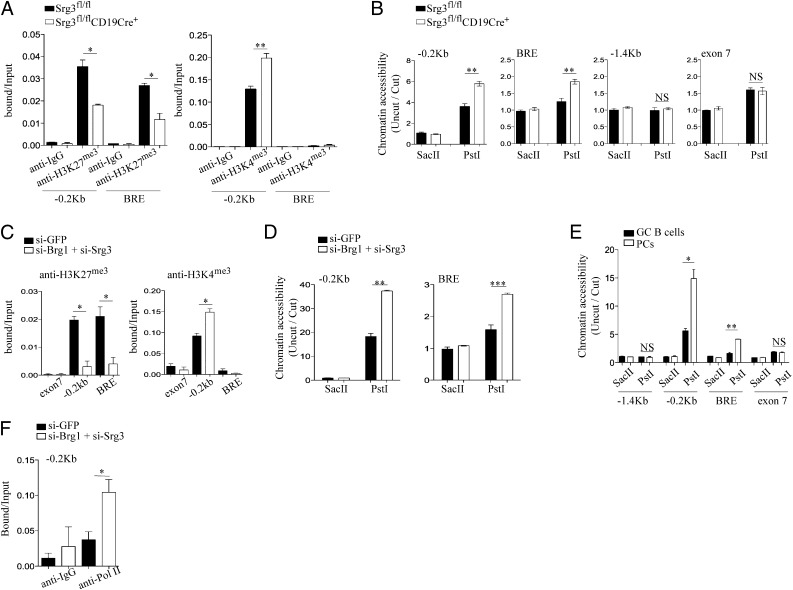
Fig. 3.
Srg3 loss impacts on the histone modification and chromatin structures at the Blimp-1 locus. (A) ChIP analysis of FO B cells obtained from Srg3fl/fl or Srg3fl/flCD19Cre+ mice with anti-isotype IgG, anti-H3K4me3, or anti-H3K27me3. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the promoter region (−0.2 kb) or BRE at the Blimp-1 locus. (B) Restriction endonuclease accessibility assay was performed on nuclei isolated from FO B cells of Srg3fl/fl or Srg3fl/flCD19Cre+ mice. Nuclei were digested with SacII or PstI and then subjected to quantitative RT-PCR analysis for the Blimp-1 locus with the indicated primers; −0.2 kb, BRE and exon 7 contain PstI recognition site, but not SacII site. The −1.4 kb does not contain both PstI and SacII recognition sites. (C) ChIP analysis of Brg1/Srg3-depleted or -intact BAL17 cells with anti-H3K27me3 or anti-H3K4me3. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the promoter region (−0.2 kb), BRE, or exon 7. (D) Restriction endonuclease accessibility with nuclei isolated from Brg1/Srg3-depleted or intact BAL17 cells. Nuclei digested with SacII or PstI were subjected to quantitative RT-PCR analysis for the promoter region and BRE. (E) Restriction endonuclease accessibility with nuclei isolated from PCs and GC B cells obtained from mice immunization with NP-KLH. Nuclei digested with SacII or PstI were subjected to quantitative RT-PCR analysis for the Blimp-1 locus with the indicated primers. (F) ChIP analysis of Brg1/Srg3-depleted or intact BAL17 cells with anti-Pol II. The precipitated DNA was analyzed by quantitative RT-PCR using the indicated primers amplifying the proximal promoter region (−0.2 kb). *P < 0.05, **P < 0.01, ***P < 0.001.