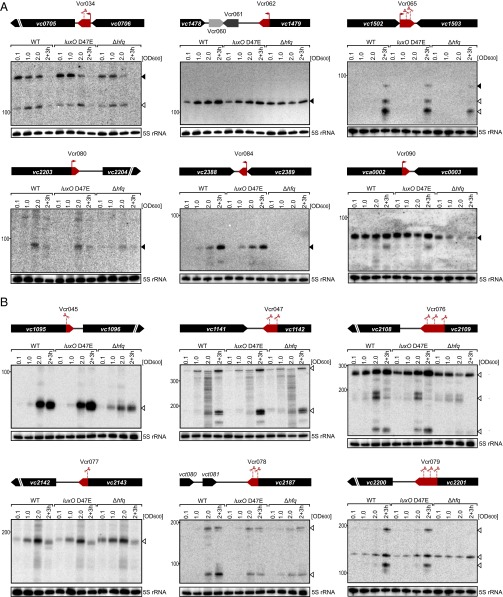
Fig. 2.
Expression analysis of 3′ UTR-derived sRNAs. (A) sRNAs with dedicated promoters. Total RNA was obtained at the designated times during growth from wild-type, luxO D47E, and Δhfq V. cholerae strains. Northern blots were probed for six 3′ UTR-derived sRNAs. The genomic locations and relative orientations are shown above the gels. Genes are shown in black; sRNAs are shown in red or gray. Arrows and scissors indicate TSSs and processing sites, respectively. Filled triangles indicate bands derived from TSSs; open triangles indicate bands derived from processing. The 5S rRNA served as the loading control. (B) sRNAs derived from transcript processing. The designations are the same as in A except that the promoters are shared between the sRNA and the mRNA. The mRNAs undergo ribonucleolytic cleavage to yield the sRNAs.