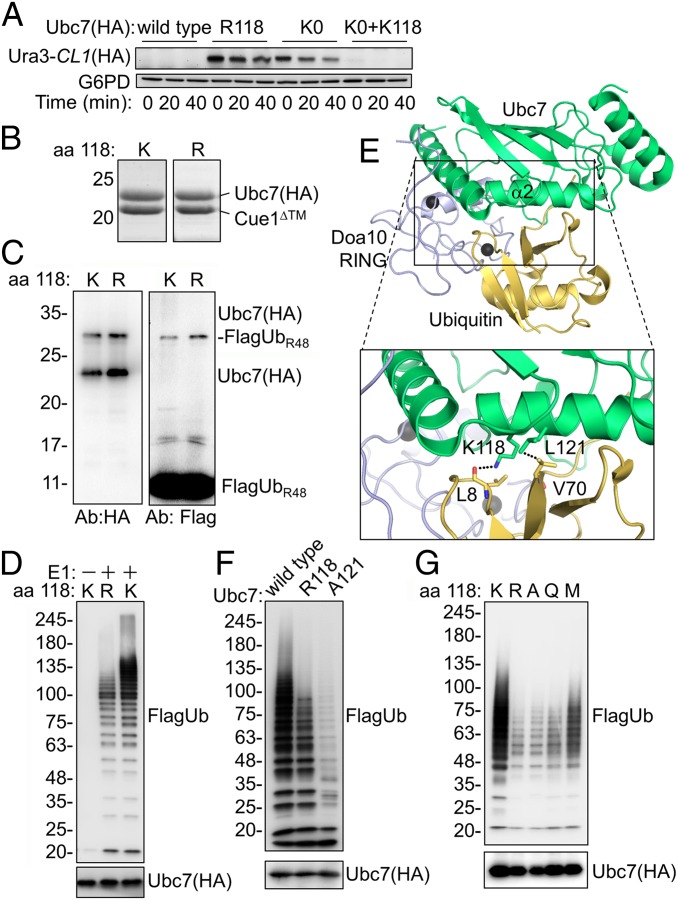
Fig. 1.
Mutations in helix α2 inhibit Ubc7 activity. (A) Ura3-CL1 degradation in cells expressing Ubc7 mutants. Cells were collected at the indicated time periods after the addition of the translation inhibitor CHX. Ura3-CL1 was visualized by immunoblotting with anti-HA Abs. Probing the blot with anti-G6PD Abs provided a loading control. (B) Coomassie staining of purified Ubc7 or Ubc7R118 shows equal amounts of copurified Cue1. Ubc7-2HA and Cue1 were expressed in bacteria from a bicistronic plasmid and subsequently were purified using a chitin-intein tag (Materials and Methods). (C) Self-monoubiquitylation of Ubc7. Purified Ubc7:Cue1 dimers were incubated with E1, ATP, and FlagUbR48 for 45 min at 30 °C. Proteins were resolved by SDS/PAGE under reducing conditions and were detected by immunoblotting with anti-Flag (Ub) or anti-HA (Ubc7) Abs. (D) PolyUb chain formation by wild-type Ubc7 or Ubc7R118. The ubiquitylation reaction was performed as described in C using FlagUb. (E) A structural model for the Doa10 RING:Ubc7∼Ub complex. The model is based on a superimposition of Ubc7 (PDB ID code 4JQU), Doa10 RING (PDB ID code 2M6M), and the RNF4 RING:UbcH5B∼Ub complex (PDB ID code 4AP4). The area within the rectangle is enlarged below. (F and G) Formation of polyUb chains by the indicated Ubc7 mutants.