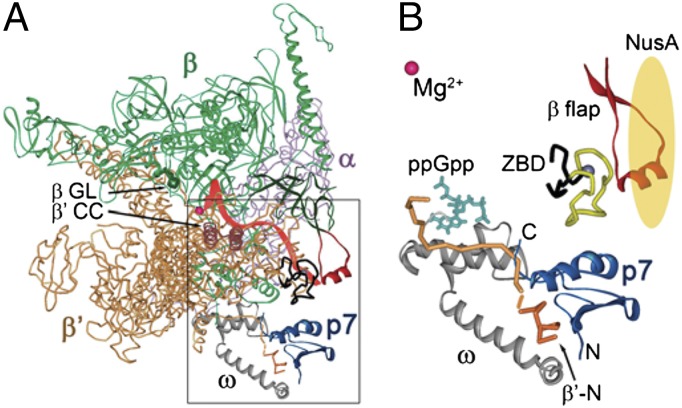
Fig. 1.
Structural model of the RNAP–p7 complex. (A) A model of the RNAP–p7 complex based on the X-ray structure of the E. coli RNAP holoenzyme 4IGC (49) and NMR structure of a complex of p7 with the N-terminal peptide of the X. oryzae β′ subunit (17). Mg2+ ion in the RNAP active center is shown as a red sphere. The σ subunit present in the original holoenzyme structure is not shown; the position of the RNA transcript (red) is drawn based on the T. thermophilus TEC structure (50). The β flap, β′ coiled-coil (β′CC), and β gate loop (βGL) elements are indicated. (B) A close-up view of the p7–RNAP interactions. P7 is shown in blue; the β′ subunit is orange, with the N-terminal peptide [residues 1–10 complexed with p7 (17)] shown in a darker tone. The ω subunit is gray; the β′ ZBD is black, with amino acid residues 70–88 deleted in this work shown in yellow; and the β flap (residues 885–921) is red. The position of ppGpp (turquoise) is superimposed from the E. coli RNAP holoenzyme ppGpp structure (4JK1) (28). NusA is schematically shown as a yellow oval.