Abstract
Background and Aims Ulmus minor has been severely affected by Dutch elm disease (DED). The introduction into Europe of the exotic Ulmus pumila, highly tolerant to DED, has resulted in it widely replacing native U. minor populations. Morphological and genetic evidence of hybridization has been reported, and thus there is a need for assessment of interspecific gene flow patterns in natural populations. This work therefore aimed at studying pollen gene flow in a remnant U. minor stand surrounded by trees of both species scattered across an agricultural landscape.
Methods All trees from a small natural stand (350 in number) and the surrounding agricultural area within a 5-km radius (89) were genotyped at six microsatellite loci. Trees were morphologically characterized as U. minor, U. pumila or intermediate phenotypes, and morphological identification was compared with Bayesian clustering of genotypes. For paternity analysis, seeds were collected in two consecutive years from 20 and 28 mother trees. Maximum likelihood paternity assignment was used to elucidate intra- and interspecific gene flow patterns.
Key Results Genetic structure analyses indicated the presence of two genetic clusters only partially matching the morphological identification. The paternity analysis results were consistent between the two consecutive years of sampling and showed high pollen immigration rates (∼0·80) and mean pollination distances (∼3 km), and a skewed distribution of reproductive success. Few intercluster pollinations and putative hybrid individuals were found.
Conclusions Pollen gene flow is not impeded in the fragmented agricultural landscape investigated. High pollen immigration and extensive pollen dispersal distances are probably counteracting the potential loss of genetic variation caused by isolation. Some evidence was also found that U. minor and U. pumila can hybridize when in sympatry. Although hybridization might have beneficial effects on both species, remnant U. minor populations represent a valuable source of genetic diversity that needs to be preserved.
Keywords: Ulmus minor, Ulmus pumila, elm, genetic diversity, hybridization, paternity analysis, fragmentation, forest remnant, conservation genetics, biological invasion, habitat degradation, long distance dispersal, plain forest.
INTRODUCTION
The paramount role played by gene flow in evolutionary processes of forest trees is often underestimated (Ellstrand, 2014). Gene flow affects how genetic variation is distributed among populations of a species, and it has been shown to influence the ability of a species to colonize new habitats and adapt to novel environments (Zalapa et al., 2010; Krutovsky et al., 2012). In conservation genetics, management strategies are strongly influenced by how gene flow affects whether most of the genetic variation is found within or among populations of a given species (Finkeldey and Ziehe, 2004; Piotti et al., 2013).
Anthropogenic changes in the environment have affected the degree of habitat fragmentation for some species, which can have a serious impact on the potential for gene flow. In general, habitat fragmentation, by reducing the size of populations and increasing the distance between them, can increase genetic drift within populations, decrease gene flow and increase mating among relatives. Such processes can lead to the loss of genetic diversity within populations and, eventually, to the fixation of alleles, even deleterious ones (Crow, 1993; Kramer et al., 2008). Aside from anthropic disturbances, natural factors, such as pathogenic attacks, climatic events and natural forest fires, can also modify the gene flow patterns characterizing forest tree populations (e.g. Shohami and Nathan, 2014). The complex interaction between the life history of a species and the current environmental conditions of its populations makes it difficult, however, to predict the actual genetic response to altered gene flow patterns without adequate information on pre- and post-disturbance gene flow estimates (Sork and Smouse, 2006; Kramer et al., 2008; Piotti, 2009; Bacles and Jump, 2011).
Anthropic and natural changes interact also in the aftermath of a pathogen attack, as in the case of Ulmus minor, the field elm (Mittempergher, 1989; Brasier, 1991, 2000). This species, which is of great ecological and cultural importance, has been repeatedly hit by Dutch elm disease (DED), caused by fungi of the genus Ophiostoma (Brasier, 2000), during the 20th century. This disease led to a drastic demographic reduction of U. minor in many European regions, including northern Italy, coupled with severe modification of its growth habit from a dominant tree to a resprouting shrub (Mittempergher, 1989). This ecological shift has allowed the congeneric species Ulmus pumila (Siberian elm) to rapidly colonize and expand in both forest and field habitats (Brunet et al., 2013). Ulmus pumila, first introduced into Europe in the 16th century (Cogolludo-Agustin et al., 2000), is a tree of considerable ruggedness and adaptability. According to a comprehensive study by Wesche et al. (2011), it is the only tree species able to withstand the harsh conditions of the semi-arid zones of the Mongolian Gobi. In addition, U. pumila is particularly prone to high hybridization rates, a characteristic that may facilitate the evolution of invasiveness (Ellstrand and Schierenbeck, 2000). Many U. pumila/Ulmus rubra hybrid trees have been identified by genetic analysis in most naturalized U. pumila populations studied across the USA (Zalapa et al., 2010), where the species was introduced as a shelter tree (Zalba and Villamil, 2002). Hybridization has been demonstrated at the morphological level among several elm species (Mittempergher and La Porta, 1991) and confirmed, by genetic methods, between U. pumila and Ulmus carpinifolia (Goodall-Copestake et al., 2005) and between U. pumila and U. minor (Cogolludo-Agustin et al., 2000; Brunet et al., 2013). Despite these warnings, the occurrence of interspecific gene flow between U. minor and U. pumila has not been investigated yet in natural populations by means of paternity analysis, which would allow parentage reconstruction between seeds and potential fathers based on their genetic compatibility (Jones et al., 2010).
Plain forests in northern Italy are nowadays extremely rare. This ecosystem has almost disappeared due to intense deforestation since the Neolithic Age, and it has been almost entirely transformed into an agricultural mosaic landscape (Marchetti, 2002). The main goal of this research was to shed light on gene flow patterns by investigating pollen movements in a natural stand of U. minor in a flood area of the Po River surrounded by trees of both species (U. minor and U. pumila) scattered in an agricultural landscape. This situation is ideal for studying intra- and interspecific gene flow. Results of paternity analysis may provide valuable information on the coexistence of native and introduced Ulmus species as a starting point for urgent conservation strategies.
MATERIALS AND METHODS
Study site and sample collection
The study area consisted of the isolated stand of Porporana (∼8 ha) and the surrounding agricultural landscape. It is located between the Po river and the village of Porporana (northern Italy, 11°28′37″ E, 44°56′03″ N; Fig. 1). To our knowledge, the Porporana stand is the last planitial forest remnant of natural origin with a high density of U. minor. Several other tree and shrub species typical of deciduous planitial forests of northern Italy are present. Most U. minor individuals are adults with an estimated age of 50–60 years. Individuals younger than 5 years are rare, probably because seedling recruitment is limited by the high tree density. This area has not been managed in the last 50 years. From a phytopathological point of view, U. minor trees in the Porporana stand are not affected by DED, with the exception of a small number of infected plants in the easternmost part of the wood (15–20 trees). Observations carried out in two consecutive years excluded the spread of infection in other areas. The Porporana stand is surrounded by arable fields for several tens of kilometres. In this agricultural area, U. pumila is more common and both species are highly scattered, with trees located several tens of metres apart.
Fig. 1.
Sampling area surrounding the relic Porporana stand. Different symbols distinguish mother trees (i.e. trees from which seeds were harvested) from all other adults trees sampled (see key). Different shadings distinguish U. minor (white) from U. pumila (black) and putative hybrid individuals (grey) according to morphological criteria (see Materials and methods).
In 2007, all elm trees with a diameter >5 cm in the Porporana stand (n = 350) and in the surrounding agricultural area within a radius of 5 km (n = 89) were sampled by collecting fresh leaves for genetic analyses and georeferenced with a GPS device. Species identification was based on the following morphological traits: (1) crown structure (mostly ascending branches in U. minor, hanging branches in U. pumila); (2) bark of younger branches (suberized and fissured in U. minor, smooth in U. pumila); (3) texture of leaf blade (matt, rough, and deeply incised by secondary venations in U. minor, shiny, smooth, with slightly incised venations in U. pumila); (4) shape of leaf edge (double indentation in U. minor, single indentation or double indentation with only the primary one marked and the secondary one sketchy in U. pumila) (Abbate et al., 2005). Trees were classified as U. minor or U. pumila if they matched at least three out of the four morphological traits described above; otherwise they were designated as hybrids. According to this morphological identification, all individuals in the Porporana stand were classified as U. minor, whereas both species and hybrids were found in the agricultural area (Fig. 1).
To elucidate the pollen dispersal dynamics, seeds were collected in 2007 and 2008 from 20 and 28 mother trees, respectively. Mother trees were chosen according to their seed production. Since seed production in the Porporana stand was low, most mother trees were located in the surrounding agricultural area (19 out of 20 in 2007, 25 out of 28 in 2008). Mother trees included U. minor (11 in 2007 and 18 in 2008), U. pumila (seven in 2007 and seven in 2008) and hybrids (two in 2007 and three in 2008).
Plant tissue collected from adult trees was stored at room temperature in 10 ml of 95 % ethanol until DNA extraction. Specimens were rehydrated at room temperature for 1 d by subsequent immersions in ethanol solutions at decreasing concentrations (70 %, 50 %, 30 %, pure water) before extraction. Harvested seeds were germinated in a greenhouse. After germination, three or four mature leaves from each seedling were used to perform DNA extraction.
DNA extraction and SSR genotyping
DNA was extracted from seedlings and adults using the DNeasy™ Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Leaf tissue was disrupted in the lysis buffer provided in the kit by the use of a TissueLyser (Qiagen) (30 Hz for 30 min). The quality and concentration of genomic DNA were assessed with an electrophoretic run (10 V/cm) on 1 % agarose gel (IBI Scientific) in 1× TAE buffer in comparison with a molecular weight standard (GeneRuler™ 1 kb DNA Ladder; Fermentas Life Sciences).
Several microsatellite markers developed for Ulmus spp. (Ulmus laevis, Whiteley et al., 2003; U. minor, Collada et al., 2004; U. rubra, Zalapa et al., 2008) were tested. After screening these markers for reproducibility, readability of amplification patterns and ability to cross-amplify between U. minor and U. pumila, two primer pairs developed by Collada et al. (2004) (Ulmi1-21, Ulmi1-165) and four loci originally developed for U. rubra by Zalapa et al. (2008) (UR123, UR138, UR158, UR188a) were retained (Supplementary Data Table S1). PCR amplification was performed in a 20-µl reaction volume containing ∼20 ng template DNA, 0·2 µm of each primer, 0·2 mm of each dNTP, 1× PCR Buffer (Promega), 2 mm MgCl2 and 0·5 U of GoTaq Flexy™ DNA Polymerase (Promega). The fluorochromes 6-FAM, HEX and TAMRA (Sigma-Genosys) were used to label the 5′ end of each forward primer. After amplification, PCR products were pooled (two triplexes, one with UR123, UR138 and Ulmi1-21 and one with UR158, UR188a and Ulmi1-165) to be run on the MegaBACE DNA sequencer. The molecular size standard ET-Rox 400™ (GE Healthcare) was used to size PCR products. Amplicons were scored using MegaBACE™ Fragment Profiler software version 1.2 (Amersham Biosciences).
Data analysis
Genetic diversity indexes
Standard genetic parameters describing genetic variation (Na, HE, HO) for adult U. minor, U. pumila and hybrid individuals were calculated with GenAlEx v. 6.5 (Peakall and Smouse, 2012). Frequencies of null alleles and the inbreeding coefficient (FIS) were estimated using the program INEst, running the individual inbreeding model (IIM) with a Gibbs sampler of 105 iterations (Chybicki and Burczyk, 2009).
Genetic structure
The Bayesian model-based clustering method implemented in STRUCTURE v.2.3.3 (Pritchard et al., 2000) was used to investigate the genetic structure in the study population. The most likely number of clusters (K) into which sampled individuals could be divided was determined using the empirical statistic ΔK, calculated using STRUCTURE HARVESTER (Evanno et al., 2005; Earl and von Holdt, 2012). STRUCTURE was run using the admixture model and correlated allele frequencies between clusters, varying K from 1 to 6. Each run consisted of 1 × 104 burn-in iterations and 5 × 104 iterations, and was replicated 20 times. Different runs for the same K value were averaged using the software CLUMPP (Jakobsson and Rosenberg, 2007) with the Greedy algorithm and 1 × 104 iterations. Individual q values quantifying the probability of belonging to one of the inferred clusters were compared with species determination based on morphological traits using Fisher’s exact test. Principal coordinates analysis was also carried out on the matrix of genetic distances among individuals in GenAlEx.
Since admixed individuals can represent either hybrids between different genetic clusters or simply genetically unrelated individuals (e.g. migrants from unsampled genetic clusters), analytical tools focused on hybrid detection were also applied. First, STRUCTURE was run again to estimate the probability that an individual was from, or had ancestry in, the inferred clusters, under the selected value of K, using prior information on populations and the options POPFLAG = 1, MIGPRIOR = 0·05 and GENSBACK = 2. Ten independent runs were done with a burn-in of 1 × 104 steps followed by 5 × 104 iterations. Second, NEWHYBRIDS (Anderson and Thompson, 2002) was used to estimate the posterior probability that each sampled individual belonged to parental classes (called P0 and P1) or hybrid categories (i.e. F1, F2, backcrosses) following two approaches: (1) no prior information about individual assignment, and (2) using individual priors corresponding to genetic clusters previously detected by STRUCTURE analysis. A burn-in of 2 × 104 steps followed by 1 × 105 iterations was used and ten independent replicates were run for each analysis.
Paternity analysis
Paternity analysis was conducted on the 386 seeds for which at least four loci were genotyped (157 in 2007 and 229 in 2008) using a maximum likelihood approach. The maximum likelihood paternity analysis was conducted on our datasets (seeds harvested in 2007 and 2008) using the software FaMoz, following a categorical allocation approach based on the assignment of paternity to the individual with the highest log of the odds (LOD) score above an estimated threshold (Gerber et al., 2003). Seeds were considered as locally pollinated if at least one compatible father with a LOD score above the threshold was found, otherwise they were classified as pollinated by external trees. The thresholds for statistical significance of LOD scores were estimated by graphically comparing (1) the distribution of LOD scores of the most likely fathers of 5 × 104 seeds randomly generated from the genotypes of local adults, representing local allele frequencies, and (2) the distribution of LOD scores of the most likely fathers of 5 × 104 seeds whose paternal genotype was randomly generated according to the allele frequencies calculated only from the paternal contribution of sampled seeds, representing allele frequencies of the background populations. The threshold was chosen at the intersection of the two distributions of LOD scores to simultaneously minimize type I error (i.e. when a seed pollinated by local pollen was not assigned to local fathers) and type II error (i.e. when a paternity is attributed to a local father whereas the true father was outside the sampling area) (Gerber et al., 2000). Thresholds were estimated for all combinations of three genotyping error values (i.e. 0·01, 0·05 and 0·10) and two inbreeding coefficient values (FIS 0 and 0·10). The e values were chosen to represent the probability of mistyping and allele dropout according to previous work (Slavov et al., 2009; Moran and Clark, 2011). To further check for the possible influence of null alleles on paternity assignment, we performed a transformation of the original dataset based on a genotype substitution procedure. In this procedure, only for loci where null allele frequencies were estimated as >0·1, homozygous genotypes were systematically changed in heterozygotes with a null allele and non-amplifying genotypes were transformed in null allele homozygotes. The procedure holds true for seed genotypes with the exception of non-amplifying seed genotypes that were not transformed if their mother trees were heterozygous. Previous studies demonstrated that this procedure significantly increases exclusion probabilities and decreases genotyping error rates when compared with other transformations (e.g. the binning procedure) (Bacles and Ennos, 2008; Piotti et al., 2012).
Maximum likelihood paternity assignments were used to estimate: (1) the pollen immigration rate from outside the study plot; (2) within-plot pollen exchanges between morphologically defined clusters and between genetically defined clusters; (3) the distribution of male reproductive success; and (4) the distribution of pollen dispersal distances. All estimates were carried out separately for the 2007 and 2008 datasets.
Pollination distances were calculated as the Euclidean distances between pollen donors and mother trees. When two (or more) pollen donors with the same LOD scores were the most likely ones, the shorter distance from the mother tree was considered to be the pollination distance. To determine whether the distribution of pollen dispersal resembled that of distances between all potential pollen donors and mother trees, we compared them using the Kolmogorov–Smirnov test (Sokal and Rohlf, 1995).
To investigate the possible origin of the paternal contribution of seeds unassigned to local fathers, we estimated their probability of belonging to STRUCTURE-inferred clusters by using the Bayesian method implemented in GeneClass2 (Rannala and Mountain, 1997; Piry et al., 2004). Pollen genotypes were determined by subtracting maternal alleles from seed genotypes and doubling them to obtain an artificial diploid pollen genotype to be analysed by an assignment test with GeneClass2. When maternal alleles could not be uniquely identified (i.e. mother and seed with the same heterozygous genotype), both alleles were included in the pollen genotype making an artificial heterozygous individual.
RESULTS
Genetic structure
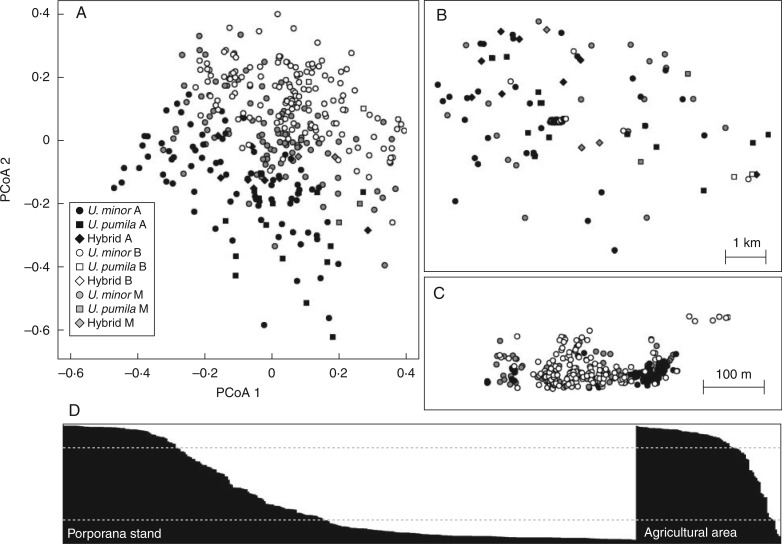
The STRUCTURE analysis, following Evanno’s method, showed the highest values of ΔK for K = 2 (Supplementary Data Fig. S1). Individuals were assigned to the two distinct genetic clusters (hereafter referred to as cluster A and cluster B) when the posterior probability of membership (q) was higher than 0·8 (Fig. 2). Individuals with lower q were all grouped in cluster M (mixed individuals). The correspondence between morphologically defined species and genetically inferred clusters was generally scarce (Fisher’s exact test, P < 0·001). Most individuals (17 out of 22) morphologically defined as U. pumila were grouped in genetic cluster A. Conversely, individuals morphologically defined as U. minor were divided between cluster A (101) and B (195), and 110 individuals were assigned to cluster M (Fig. 2; for raw data see Supplementary Data Table S2). In the Porporana stand, trees were mainly classified as B (191/350) whereas cluster A was less represented (69/350, Fig. 2C). On the contrary, in the surrounding agricultural area most individuals were classified as A or M, whereas only six individuals were assigned to cluster B (Fig. 2B). Principal coordinates analysis analysis confirmed results from Bayesian clustering (Fig. 2A). The first two principal coordinates, which explained 49 % of the total genetic variation, showed a clear distinction between morphologically identified U. minor trees located in the Porporana stand and those in the surrounding area (open and black circles, respectively, in Fig. 2B and C). In the principal coordinates analysis plot, the putative morphological hybrids (diamonds in Fig. 2A) are all located in an intermediate position between genetic cluster A (open symbols) and B (black symbols in Fig. 2A).
Fig. 2.
Results of cluster analyses. (A) Principal coordinates analysis scatterplot, with different shadings indicating different clusters according to STRUCTURE analysis and different symbols indicating morphological identification. (B) The same shading and symbol representation plotted on the geographical position of individuals, zooming in on the central Porporana stand (C). (D) Distribution of q values from STRUCTURE analysis for the Porporana stand (on the left) and the surrounding agricultural area (on the right). The black and white portions of each bar are the probabilities of belonging to cluster A and B, respectively. Grey dotted lines represent 0·8 thresholds applied for assignment to clusters A and B.
The probability that an individual had ancestry in the inferred clusters A and B was estimated by running STRUCTURE again with the GENSBACK option on. Under the prior of the two clusters, all admixed individuals (cluster M) derived half of their genetic setup from each cluster. In no cases did M trees show ancestry from either cluster A or cluster B (Supplementary Data Table S3).
The presence of hybrids was further investigated using the software NEWHYBRIDS with or without prior information on the genetic assignment. In both cases, there was general agreement among likelihood distributions of ten independent runs that gave highly comparable results. Therefore, results from a single representative run for each analysis, choosing a threshold for q values at 0·80, are presented in Supplementary Table S4. In the analysis using the assignment to A and B clusters by the use of STRUCTURE as prior information, only one individual from the M cluster was classified as a hybrid, whereas the others were assigned to parental class P1 or unassigned. The low number of individuals classified as hybrids was confirmed by the analysis without prior information. The majority of individuals classified as admixed using STRUCTURE (cluster M) fell into the P1 category, with 16 unassigned and 9 individuals out of 116 classified as hybrids (Supplementary Data Table S4).
Paternity analysis
The markers used were highly polymorphic (mean Na = 10·833, mean HE = 0·69) and provided good resolution in terms of exclusion probability (EP = 0·987, Supplementary Data Table S5). The transformation applied to original datasets did not reveal substantial changes in gene flow estimates. Varying paternity analysis settings (increasing FIS and e), we obtained consistently high gene flow estimates, ranging from 0·59 to 0·89 when considering the two years together (Supplementary Data Table S6). In the following, we chose to use the paternity assignments obtained from the intermediate scenario based on the original dataset, with FIS = 0·10 and e = 0·05.
Analysing paternity assignments from this scenario, we found that 33 seeds out of the 157 sampled (21 %) in 2007 and 53 seeds out of the 229 sampled (23 %) in 2008 were compatible with at least one pollen donor inside the study area. Conversely, 124 and 176 seeds in 2007 and 2008, respectively, had no compatible pollen donor inside the study plot, giving a pollen immigration rate from outside the sampling area of 0·79 and 0·77, respectively. Among locally pollinated seeds, one in 2007 and four in 2008 were self-pollinated. The pollen immigration rate differed among mother trees, ranging from 0·5 to 1·0 in 2007 and from 0·2 to 1·0 in 2008 (Supplementary Data Table S7), although a higher number of seeds per mother tree would have been required to obtain precise estimates of pollen immigration at the mother tree level. The pollen immigration rate was similar among genetically inferred clusters (Table 1).
Table 1.
Numbers of pollination events detected among and within genetic clusters (from STRUCTURE analysis) in 2007 and 2008
| Year | From A | % | From B | % | From M | % | From outside | % | |
|---|---|---|---|---|---|---|---|---|---|
| 2007 | To A | 10 | 11 | 4 | 4 | 9 | 10 | 69 | 75 |
| To B | 1 | 7 | 2 | 14 | 1 | 7 | 10 | 71 | |
| To M | 0 | 0 | 4 | 8 | 2 | 4 | 45 | 88 | |
| 2008 | To A | 21 | 20 | 1 | 0 | 10 | 10 | 72 | 69 |
| To B | 0 | 0 | 0 | 0 | 1 | 6 | 15 | 94 | |
| To M | 5 | 5 | 3 | 3 | 12 | 11 | 89 | 81 |
Despite the higher number of potential pollen donors belonging to cluster B, which represents mainly individuals from the Porporana stand, effective pollination events from B individuals to A and M mother trees were infrequent. On the other hand, most pollinations from A individuals were directed towards A mother trees. Five cases of intercluster pollinations were recorded in 2007 (one from A to B and four from B to A) and one from B to A in 2008 (Table 1).
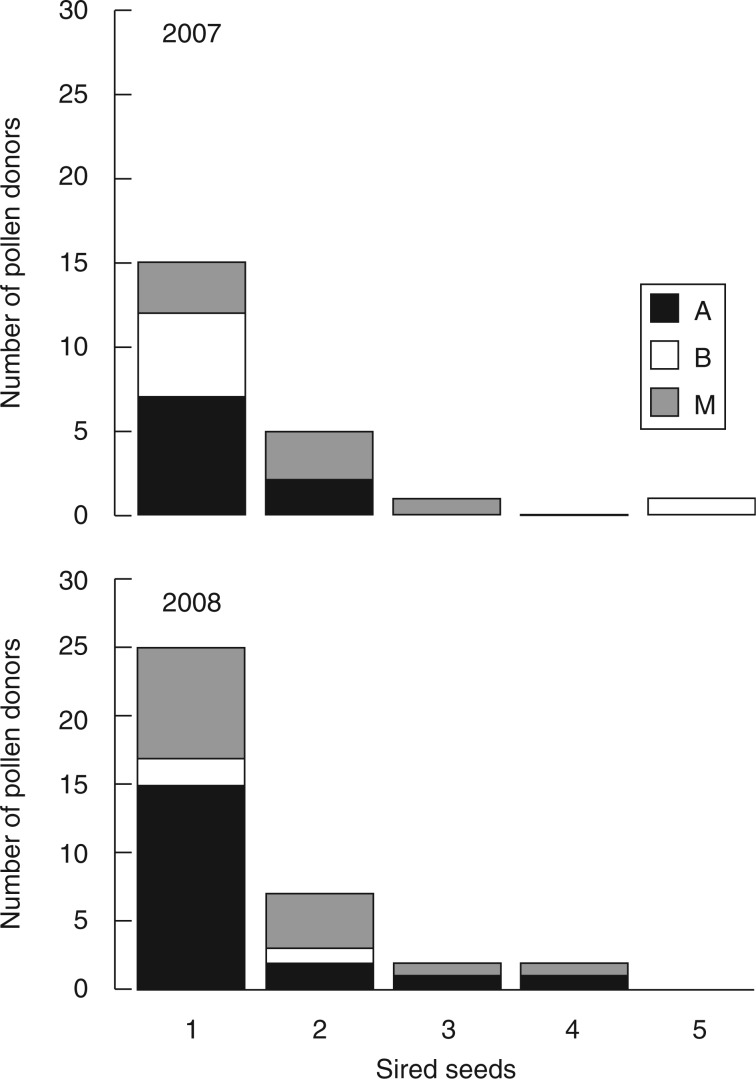
Few local individuals were involved in the pollination process. Only 22 and 36 individuals (out of 439) acted as successful pollen donors in 2007 and 2008, respectively. The distribution of male reproductive success (i.e. the number of sired seeds per potential pollen donor) was skewed, with few individuals involved in the majority of local pollination events. Seven adults were involved in 55 % of local pollinations in 2007, and 11 in 53 % of local pollinations in 2008. Most successful pollen donors were individuals belonging to clusters A and M (Fig. 3). The higher involvement of cluster A-like pollen donors to background pollination dynamics in the study plot was confirmed by the cluster assignment test carried out on paternally unassigned pollen genotypes using GeneClass2. Most unassigned pollen genotypes were compatible with the genetic composition of cluster A (Supplementary Data Fig. S2).
Fig. 3.
Number of pollen donors from different clusters (according to STRUCTURE analysis) in relation to their reproductive success in terms of the number of sired seeds in 2007 and 2008.
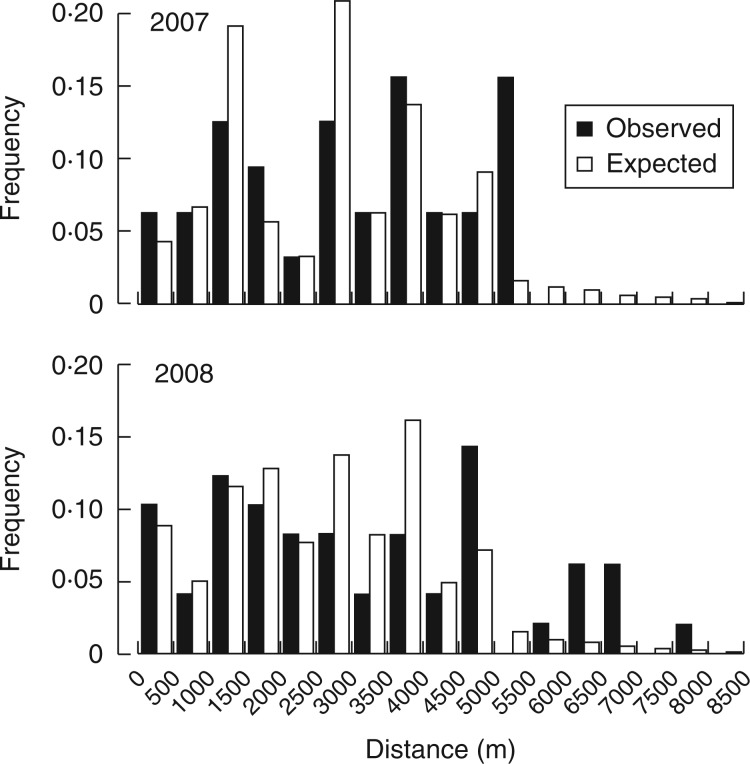
Mean (± s.d.) pollination distances were high and comparable between years (2·86 ± 1·66 km in 2007 and 2·93 ± 2·14 km in 2008) (Fig. 4). The distribution of pollen dispersal distances in 2007 was not different from the expected distribution considering the distances among all possible pollen donors and mother trees (Kolmogorov–Smirnov test: D = 0·14, P = 0·51), whereas in 2008 there was a lack of intermediate distances compensated for by an excess of pollination events exceeding 5 km (D = 0·21, P < 0·05). The maximum pollination distances recorded were extremely high in both years (5·42 km in 2007 and 7·89 km in 2008) (Fig. 4).
Fig. 4.
Distribution of observed pollen dispersal distances (black bars) and distribution of all distances between potential pollen donors and maternal trees (open bars) in 2007 and 2008.
DISCUSSION
In this work, we investigated pollen dispersal patterns and population structure sampling all adult elm trees within a wide area (5 km radius = ∼80 km2) centred on a dense relic stand of U. minor surrounded by an agricultural area where individuals of the two species present (U. minor and U. pumila) have a scattered distribution. By using paternity analysis, we estimated the pollen immigration rate from outside the sampling area and the characteristics of local pollination patterns, as well as pollen exchanges between the two genetic clusters detected by Bayesian clustering, in two consecutive reproductive seasons. We found that pollen immigration rates were high (∼0·6–0·9) and comparable between the two reproductive seasons investigated. Despite the scattered distribution of individuals inside the sampling area, the analysis of within-population pollination patterns showed that pollen dispersal distances were extremely large (up to 8 km), confirming that the high pollen dispersal capabilities of the two species can maintain the genetic connectivity among isolated individuals and stands. Furthermore, our findings about the presence of few intercluster pollinations and hybrid individuals suggest that hybridization, albeit rare, has been occurring, supporting previous evidence for low mating barriers between U. minor and U. pumila.
Pollen gene flow in a fragmented landscape
The general picture emerging from our study about pollen dispersal patterns in the two Ulmus species investigated is characterized by high pollen immigration and extremely low self-pollination (0·6 % in 2007 and 0·8 % in 2008). This confirms previous results obtained at a smaller spatial scale in other Ulmus species. Nielsen and Kjær (2010a, b) investigated pollen dispersal patterns in U. laevis and Ulmus glabra, showing how both species are characterized by high outcrossing rates and the absence of self-pollination. In U. laevis, they found dispersal events up to 1 km, and such long pollination events were also tracked among isolated U. glabra trees. Interestingly, when comparing gene flow patterns between continuous forest and open landscape conditions, Nielsen and Kjær (2010b) found that mean pollen dispersal was ∼100 m in the continuous forest and much greater among scattered individuals. As in the present study, this shows that scattered trees may act as important bridges for pollen exchange connecting remnant stands. The role of scattered trees in connecting fragmented populations has been highlighted previously, mainly in insect- and animal-pollinated trees (Levin, 1995; Aldrich and Hamrick, 1998; White et al., 2002; Kamm et al., 2009). Our study strengthens the increasing evidence that pollination can be effective at the landscape level also in fragmented wind-pollinated temperate species (Bacles and Ennos, 2008; Robledo-Arnuncio, 2011; Leonardi et al., 2012). On the other hand, information on seed gene flow is lacking for Ulmus spp. Bläß et al. (2010) reported that elm seeds can be transported only occasionally by a distance of >100 m and results on the spatial genetic structure of natural elm populations suggest limited transport of seeds by wind (Nielsen and Kjær, 2010a). Further investigation is needed to understand the relative impacts of fragmentation on pollen and seed dispersal.
Gene flow at interspecific level
Another goal of our work was to assess the amount of cross-pollination between U. minor and U. pumila and the consequent formation of hybrids. In this context, it is worth noting that the ambiguity of the morphological identification was stressed in previous work on Ulmus species (e.g. Zalapa et al., 2010). In our case, this was reflected in the poor agreement with the classification according to genetic criteria (i.e. membership coefficients of STRUCTURE), although a higher number and/or different classes of markers might improve the discrimination between the two species. However, substantial agreement between the morphological and genetic classifications was found for individuals that were morphologically identified as U. pumila (mainly belonging to cluster A) and for U. minor individuals from the Porporana stand (mainly belonging to cluster B).
The presence of few putative hybrids among adult individuals, along with some inter-cluster pollination events, adds further evidence about the occurrence of hybridization between Ulmus species (Cogolludo-Agustin et al., 2000; Goodall-Copestake et al., 2005; Zalapa et al., 2010; Brunet et al., 2013). Some of the main key factors promoting possible hybridization are the weak barriers to cross-fertilization characterizing the genus Ulmus (Mittempergher and La Porta, 1991) and the observation that anthropic alteration of environments could facilitate hybridization in forest trees (Hoban et al., 2012). Unlike previous hybridization studies on U. rubra/U. pumila (Zalapa et al., 2009) and U. minor/U. pumila (Brunet et al., 2013), in which backcrosses were biased towards U. pumila, our data show a more balanced situation. However, the few local pollination events detected towards mother trees from cluster M (six in 2007 and 20 in 2008) suggest the need for caution about this issue.
Previous studies suggested a possible role played by DED in the recent biological history of sympatric U. minor and U. pumila populations (Solla et al., 2005; Brunet et al., 2013). They hypothesized that the native species U. minor, heavily depleted by the effect of DED epidemics, has introgressed some of the U. pumila’s DED resistance alleles. On the other hand, traits introgressed from autochthonous to allochthonous species could made them fitter in the new environment and, eventually, better colonizers (Ellstrand and Schierenbeck, 2000; Sakai et al., 2001; Hegde et al., 2006). Such is the scenario depicted for U. pumila/U. rubra by Zalapa et al. (2009). They showed that hybridization between U. pumila and the autochthonous American species U. rubra is strongly unbalanced towards the former, likely enhancing its fitness in the new environment rather than introgressing DED resistance into U. rubra. However, the high pollen immigration from outside and the few intercluster pollination events detected in our study do not allow us to speculate further on such evolutionary dynamics.
Conservation issues
Previous studies have demonstrated that gene flow can be restricted in isolated populations of forest trees (Bittencourt and Sebben, 2007; Hanaoka et al., 2007; Sebben et al., 2010). This lack of genetic connectivity would exacerbate the effect of genetic drift, speeding up the loss of genetic variation in forest remnants. However, there is increasing evidence that spatially isolated trees and forest fragments are not necessarily genetically isolated, often receiving sufficient gene flow, in particular via pollen, to counteract the detrimental effects of fragmentation (Aldrich and Hamrick, 1998; Kamm et al., 2009; Buschbom et al., 2011; Robledo-Arnuncio, 2011). One possible explanation is that high local densities can impede long-distance dispersal because of high local pollen availability and by changing the aerodynamic properties of propagule dispersal (Knapp et al., 2001; Hardy, 2009; Piotti et al., 2012). Shohami and Nathan (2014) went further, demonstrating how disturbances that reduce, even drastically, local density can facilitate dispersal across extensive distances. In this work, based on a highly fragmented agricultural landscape, we found much longer pollen dispersal distances than previously found in Ulmus spp. and massive pollen immigration from outside. We can thus suggest than gene flow via pollen is not a limiting factor at this degree of fragmentation. Therefore, our results on gene flow patterns, together with the observation of scarce regeneration within the Porporana stand, confirm the general view that the genetic consequences of fragmentation can often be more delayed than ecological ones [i.e. recruitment failure due to pollen limitation and reduction of optimal habitat (Kramer et al., 2008)]. This calls for increasing effort to couple ecological and genetic data to detect early signs of the fragmentation process in the short-term evolutionary dynamics of forest remnants.
Although U. minor is not considered an endangered species despite DED pandemics, it is unequivocally evident that the effects of this disease, together with the sympatric occurrence of the congeneric, invasive U. pumila, has modified the original ecological and genetic status of the European species (Mittempergher, 1989; Brunet et al., 2013). First, alteration of its growth habit changed the ecological role of the species itself; second, a dramatic demographic reduction exposed U. minor to the possible consequences of a fragmented distribution. Our results provide important information for realizing effective conservation strategies for the Porporana stand and surrounding areas. Particular care should be taken in preserving forest remnants with U. minor, which constitute an important genetic archive of the pre-DED species’ genetic pool. In harvesting seeds for afforestation programmes, relict ancient woods should be preferentially chosen.
We have to stress that this was preliminary research aimed at describing gene flow patterns and the genetic structure of a population in a particular ecological context. To investigate the genetic consequences of the coexistence of native and introduced Ulmus species, it is necessary to acquire a more comprehensive picture based on the analysis of a large set of markers, which should include candidate genes with potential adaptive value. Another aspect deserving further attention will be the monitoring of gene flow patterns in recently renaturalized areas during the transition from a young reafforestation stand into mature wood.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: ΔK values from 20 replicates of the STRUCTURE analysis calculated using STRUCTURE HARVESTER, for K from 1 to 8. Figure S2: probabilities of unassigned pollen genotypes belonging to STRUCTURE-inferred clusters estimated by GeneClass2. Table S1: characteristics of the marker set. Table S2: correspondence between morphological classification and genetic clustering. Table S3: hybrid detection analysis with STRUCTURE. Table S4. Hybrid detection analysis with NEWHYBRIDS. Table S5: genetic diversity parameters locus by locus for the entire dataset and the Porporana stand only, calculated using GenAlEx. Table S6: comparison of pollen immigration estimates obtained by varying FaMoz settings for genotyping error values and inbreeding coefficient values on the original and transformed datasets. Table S7: pollen immigration rate for each mother tree in 2007 and 2008.
ACKNOWLEDGEMENTS
This work was partially funded by MiPAAF, Corpo Forestale dello Stato (to B.B., L.Z., L.G. and F.G.) and by project FAR 2012 of the University of Insubria (to G.B.). We are grateful to Genexpress Laboratory (University of Florence, Italy) for help in the genetic analysis. We also thank the editor and three anonymous reviewers for helpful comments on an earlier version of the manuscript.
LITERATURE CITED
- Abbate G, Alessandrini A, Blasi C, Conti F. 2005. An annotated checklist of the Italian vascular flora. Rome: Palombi Editori. [Google Scholar]
- Aldrich PR, Hamrick JL. 1998. Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 281: 103–105. [DOI] [PubMed] [Google Scholar]
- Anderson EC, Thompson EA. 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160: 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacles CFE, Ennos RA. 2008. Paternity analysis of pollen-mediated gene flow for Fraxinus excelsior L. in a chronically fragmented landscape. Heredity 101: 368–380. [DOI] [PubMed] [Google Scholar]
- Bacles CFE, Jump AS. 2011. Taking a tree’s perspective on forest fragmentation genetics. Trends in Plant Science 16: 13–18. [DOI] [PubMed] [Google Scholar]
- Bittencourt JVM, Sebbenn AM. 2007. Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99: 580–591. [DOI] [PubMed] [Google Scholar]
- Bläß C, Ronnenberg K, Tackenberg O, Hensen I, Wesche K. 2010. The relative importance of different seed dispersal modes in dry Mongolian rangelands. Journal of Arid Environments 74: 991–997. [Google Scholar]
- Brasier CM. 1991. Ophiostoma novo-ulmi sp. nov., causative agent of current Dutch elm disease pandemics. Mycopathologia 115: 151–161. [Google Scholar]
- Brasier CM. 2000. Intercontinental spread and continuing evolution of the Dutch elm disease pathogens. In: Dunn C. ed. The elms: breeding, conservation and disease management. Dordrecht: Kluwer, 61–72. [Google Scholar]
- Brunet J, Zalapa JE, Pecori F, Santini A. 2013. Hybridization and introgression between the exotic Siberian elm, Ulmus pumila, and the native field elm, U. minor, in Italy. Biological Invasions 15: 2717–2730. [Google Scholar]
- Buschbom J, Yanbaev Y, Degen B. 2011. Efficient long-distance gene flow into an isolated relict oak stand. Journal of Heredity 102: 464–472. [DOI] [PubMed] [Google Scholar]
- Chybicki IJ, Burczyk J. 2009. Simultaneous estimation of null alleles and inbreeding coefficients. Journal of Heredity 100: 106–113. [DOI] [PubMed] [Google Scholar]
- Cogolludo-Agustin MA, Agundez D, Gil L. 2000. Identification of native and hybrid elms in Spain using isozyme gene markers. Heredity 85: 157–166. [DOI] [PubMed] [Google Scholar]
- Collada C, Fuentes-Utrilla P, Gil L, Cervera MT. 2004. Characterization of microsatellite loci in Ulmus minor Miller and cross-amplification in U. glabra Hudson and U. laevis Pall. Molecular Ecology Notes 4: 731–732. [Google Scholar]
- Crow JF. 1993. Mutation, mean fitness, and genetic load. In: Futuyma D, Antonovics J. eds. Oxford surveys in evolutionary biology, Vol. 9 Oxford: Oxford University Press, 3–42. [Google Scholar]
- Earl DA, von Holdt BM. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetic Resources 4: 359–361. [Google Scholar]
- Ellstrand NC. 2014. Is gene flow the most important evolutionary force in plants? American Journal of Botany 101: 737–753. [DOI] [PubMed] [Google Scholar]
- Ellstrand NC, Schierenbeck KA. 2000. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the USA 97: 7043–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Finkeldey R, Ziehe M. 2004. Genetic implications of silvicultural regimes. Forest Ecology and Management 197: 231–244. [Google Scholar]
- Gerber S, Mariette S, Streiff R, Bodenes C, Kremer A. 2000. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Molecular Ecology 9: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Gerber S, Chabrier P, Kremer A. 2003. FAMOZ: a software for parentage analysis using dominant, codominant and uniparentally inherited markers. Molecular Ecology Notes 3: 479–481. [Google Scholar]
- Goodall-Copestake WP, Hollingsworth ML, Jenkins GI, Collin E. 2005. Molecular markers and ex situ conservation of the European elms (Ulmus spp.). Biological Conservation 122: 537–546. [Google Scholar]
- Hanaoka S, Yuzurihara J, Asuka Y, et al. 2007. Pollen-mediated gene flow in a small, fragmented natural population of Fagus crenata. Canadian Journal of Botany 85: 404–413. [Google Scholar]
- Hardy OJ. 2009. How fat is the tail? Heredity 103: 437–438. [DOI] [PubMed] [Google Scholar]
- Hegde SG, Nason JD, Clegg JM, Ellstrand NC. 2006. The evolution of California’s wild radish has resulted in the extinction of its progenitors. Evolution 60: 1187–1197. [PubMed] [Google Scholar]
- Hoban SM, McCleary TS, Schlarbaum SE, Anagnostakis SL, Romero-Severson J. 2012. Human-impacted landscapes facilitate hybridization between a native and an introduced tree. Evolutionary Ecology 5: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jones AG, Small CM, Paczolt KA, Ratterman NL. 2010. A practical guide to methods of parentage analysis. Molecular Ecology Resources 10: 6–30. [DOI] [PubMed] [Google Scholar]
- Kamm U, Rotach P, Gugerli F, Siroky M, Edwards P, Holderegger R. 2009. Frequent long-distance gene flow in a rare temperate forest tree (Sorbus domestica) at the landscape scale. Heredity 103: 476–482. [DOI] [PubMed] [Google Scholar]
- Knapp EE, Goedde MA, Rice KJ. 2001. Pollen-limited reproduction in blue oak: implications for wind pollination in fragmented populations. Oecologia 128: 48–55. [DOI] [PubMed] [Google Scholar]
- Kramer AT, Ison JL, Ashley MV, Howe HF. 2008. The paradox of forest fragmentation genetics. Conservation Biology 22: 878–885. [DOI] [PubMed] [Google Scholar]
- Krutovsky KV, BurczyK J, Chybicki I, Finkeldey R, Pyhäjärvi T, Robledo-Arnuncio JJ. 2012. Gene flow, spatial structure, local adaptation, and assisted migration in trees. In: Schnell RJ, Priyadarshan PM. eds. Genomics of tree crops. New York: Springer, 71–116. [Google Scholar]
- Leonardi S, Piovani P, Scalfi M, Piotti A, Giannini R, Menozzi P. 2012. Effect of habitat fragmentation on the genetic diversity and structure of peripheral populations of beech in Central Italy. Journal of Heredity 103: 408–417. [DOI] [PubMed] [Google Scholar]
- Levin DA. 1995. Plant outliers: an ecogenetic perspective. American Naturalist 145: 109–118. [Google Scholar]
- Marchetti M. 2002. Environmental changes in the central Po Plain (northern Italy) due to fluvial modifications and anthropogenic activities. Geomorphology 44: 361–373. [Google Scholar]
- Mittempergher L. 1989. Il declino dell’olmo: da latifoglia nobile a cespuglio. Annali Accademia Italiana Scienze Forestali 38: 585–609. [Google Scholar]
- Mittempergher L, La Porta N. 1991. Hybridization studies in the Eurasian species of elm (Ulmus spp.). Silvae Genetica 40: 237–243. [Google Scholar]
- Moran EV, Clark JS. 2011. Estimating seed and pollen movement in a monoecious plant: a hierarchical Bayesian approach integrating genetic and ecological data. Molecular Ecology 20: 1248–1262. [DOI] [PubMed] [Google Scholar]
- Nielsen LR, Kjær ED. 2010a. Fine-scale gene flow and genetic structure in a relic Ulmus laevis population at its northern range. Tree Genetics & Genomes 6: 643–649. [Google Scholar]
- Nielsen LR, Kjær ED. 2010b. Gene flow and mating patterns in individuals of wych elm (Ulmus glabra) in forest and open land after the influence of Dutch elm disease. Conservation Genetics 11: 257–268. [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotti A. 2009. The genetic consequences of habitat fragmentation: the case of forests. iForest 2: 75–76. [Google Scholar]
- Piotti A, Leonardi S, Buiteveld J, et al. 2012. Comparison of pollen gene flow among four European beech (Fagus sylvatica L.) populations characterized by different management regimes. Heredity 108: 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotti A, Leonardi S, Heuertz M, et al. 2013. Within-population genetic structure in beech (Fagus sylvatica L.) stands characterized by different disturbance histories: does forest management simplify population substructure? PLoS ONE 8: e73391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L, Estoup A. 2004. GeneClass2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity 95: 536–539. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly O. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Mountain JL. 1997. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences of the USA 94: 9197–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo-Arnuncio JJ. 2011. Wind pollination over mesoscale distances: an investigation with Scots pine. New Phytologist 190: 222–233. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. 2001. The population biology of invasive species. Annual Review of Ecology, Evolution and Systematics 32: 305–332. [Google Scholar]
- Sebbenn AM, Carvalho ACM, Freitas, et al. 2010. Low levels of realized seed and pollen gene flow and strong spatial genetic structure in a small, isolated and fragmented population of the tropical tree Copaifera langsdorffii Desf. Heredity 106: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohami D, Nathan R. 2014. Fire-induced population reduction and landscape opening increases gene flow via pollen dispersal in Pinus halepensis. Molecular Ecology 23: 70–81. [DOI] [PubMed] [Google Scholar]
- Slavov GT, Leonardi S, Burczyk J, Adams WT, Strauss SH, DiFazio SP. 2009. Extensive pollen flow in two ecologically contrasting populations of Populus trichocarpa. Molecular Ecology 18: 357–373. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. 1995. Biometry: principles and practices of statistics in biological research, 3rd edn New York: W. H. Freeman. [Google Scholar]
- Solla A, Bohnens J, Collin E, et al. 2005. Screening European elms for resistance to Ophiostoma novo-ulmi. Forest Science 51: 134–141. [Google Scholar]
- Sork VL, Smouse PE. 2006. Genetic analysis of landscape connectivity in tree populations. Landscape Ecology 21: 821–836. [Google Scholar]
- Wesche K, Walther D, von Wehrden H, Hensen I. 2011. Trees in the desert: reproduction and genetic structure of fragmented Ulmus pumila forests in Mongolian drylands. Flora 206: 91–99. [Google Scholar]
- White GM, Boshier DH, Powell W. 2002. Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proceedings of the National Academy of Sciences of the USA 99: 2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley RE, Black-Samuelsson S, Clapham D. 2003. Development of microsatellite markers for the European white elm (Ulmus laevis Pall.) and cross-species amplification within the genus Ulmus. Molecular Ecology Notes 3: 598–600. [Google Scholar]
- Zalapa JE, Brunet J, Guries RP. 2008. Isolation and characterization of microsatellite markers for red elm (Ulmus rubra Muhl.) and cross-species amplification with Siberian elm (Ulmus pumila L.). Molecular Ecology Resources 8: 109–112. [DOI] [PubMed] [Google Scholar]
- Zalapa JE, Brunet J, Guries RP. 2009. Patterns of hybridization and introgression between invasive Ulmus pumila (Ulmaceae) and native U . rubra. American Journal of Botany 96: 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zalapa JE, Brunet J, Guries RP. 2010. The extent of hybridization and its impact on the genetic diversity and population structure of an invasive tree, Ulmus pumila (Ulmaceae). Evolutionary Applications 3: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalba SM, Villamil CB. 2002. Woody plant invasion in relictual grasslands. Biological Invasions 4: 55–72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.