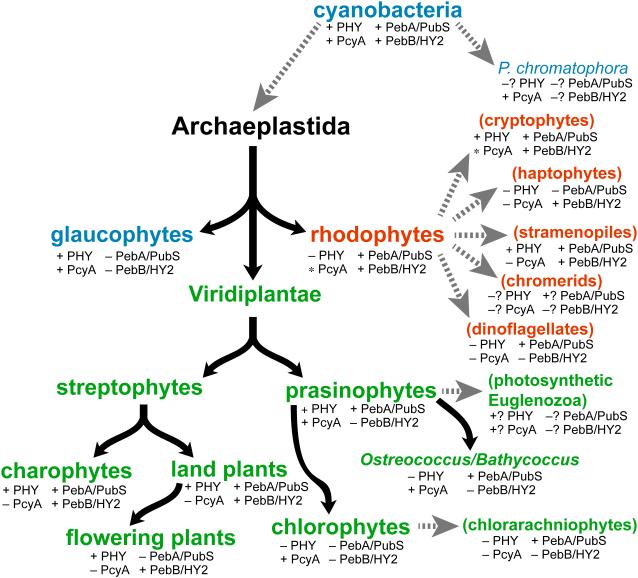
Figure 4.
Distribution of phytochromes and FDBRs in eukaryotic algae. Evolution of primary algae (Archaeplastidae) is shown, with glaucophyte (blue), rhodophyte (red) and Viridiplantae (green) lineages. We define subsequent evolution of the Viridiplantae with an initial split into streptophytes and prasinophytes. The streptophytes comprise modern charophytes and land plants (embryophytes). Modern charophyte and prasinophyte algae are paraphyletic, with land plants and chlorophyte algae descending from charophytes and prasinophytes, respectively (Worden et al., 2009; Timme et al., 2012; Duanmu et al., 2014). Subsequent endosymbioses (dashed grey arrows) gave rise to secondary algae (parentheses), which are color-coded to indicate the Archaeplastida lineage that was assimilated. Tertiary endosymbioses of diatoms by dinoflagellates are not shown. Distribution of phytochromes was assessed by performing BLAST searches of genomic and transcriptomic data (with default parameters) using the cyanobacterial phytochrome Cph1 as a query sequence. The presence of FDBRs was assessed using a similar strategy, with PcyA from Anabaena sp. strain PCC 7120 and PebA and PebB from Nostoc punctiforme as query sequences. FDBRs were assigned to the PcyA, PebA, or PebB lineage based on BLAST scores with the three query sequences, an approach that provided both unambiguous assignment of algal FDBRs and recovery of the three lineages detected previously (Chen et al., 2012). The presence of PcyA only in certain divisions within rhodophytes and cryptophytes is indicated by the asterisk (e.g., presence of PcyA in the cryptophyte genus Hemiselmis). Question marks indicate groups for which complete draft genomes and/or >3 transcriptomic datasets are not yet available.