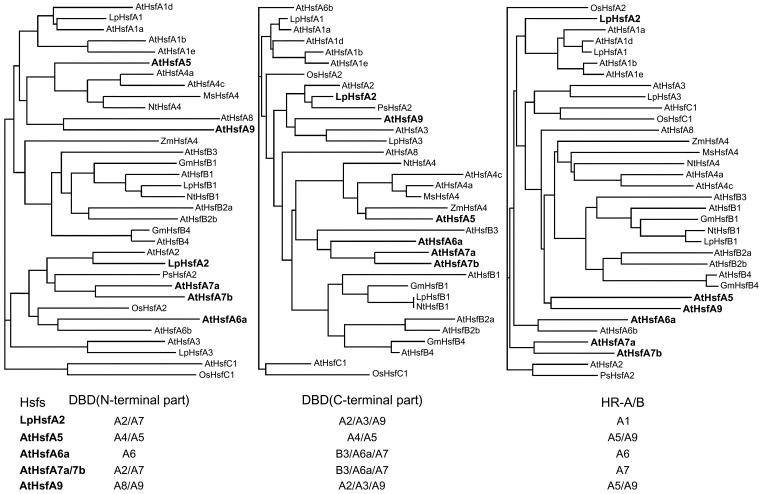
Fig 4.
Clustal analysis of the phylogenetic relationship based on the comparison of sequences of different parts of the N-terminal half of plant Hsfs. Three different conserved parts were used to create the phylogenetic trees: (1) the N-terminal part of the DBD until the position of the intron, (2) the C-terminal part of the DBD, and (3) the HR-A/B region, including 2 heptad repeats of HR-A, the insert, and HR-B (see borders defined in Table 2). Most Hsfs are found in identical groups irrespective of the sequence parts used for the analysis. However, a few Hsfs marked by boldfaced letters change their positions (see also the summary given at the bottom of the figure). We assume that, similar to the situation in the present mammalian Hsf genes, these 3 parts were separated by introns in the ancient plant Hsf precursor gene, ie, exon shuffling could have generated mosaic Hsfs. For AtHsfs A7a and A7b, the following situation is envisaged. The N-terminal part of the DBD and the HR-A/B region are derived from the putative A2/A7 precursor gene, whereas the C-terminal part of the DBD stems are from the B3/A7/A6a precursor gene