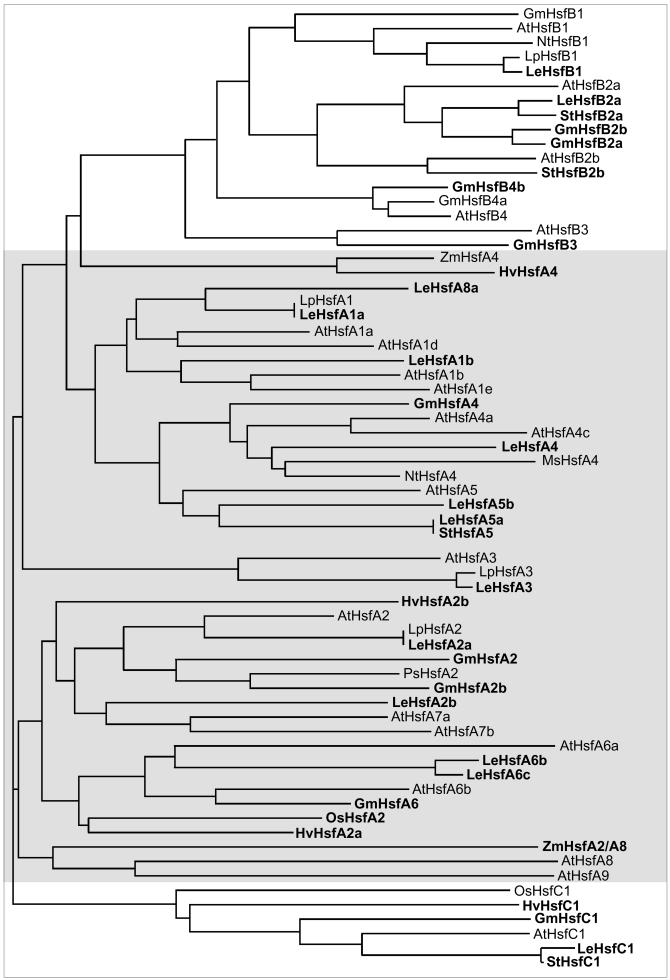
Fig 5.
Extended version of phylogenetic relationships of plant Hsfs. To emphasize the complexity of the plant Hsf family in general, an extended version of the phylogenetic tree was created by Clustal analysis based on the N-terminal parts of the DBD of those Hsfs contained in Fig 3 plus a considerable number of additional partial clones derived from the database, mostly from EST libraries of tomato (Lycopersicon esculentum), potato (Solanum tuberosum), soybean (Glycine max), and barley (Hordeum vulgare). For other abbreviations see Fig 3 and Table 1. New entries in the figure are marked by boldfaced letters. Information was derived from the following ESTs and accession numbers: L esculentum HsfA1a (AW933448, AW399336, AW223123), HsfA1b (BE354387), HsfA2a (AW034874), HsfA2b (AW930998), HsfA3 (BE433610, AI895834, AW034135, AW035854, AW035844, AW030642, AW033013; all ESTs represent incompletely spliced messenger RNAs), HsfA4 (AW038959, AW933529), HsfA5a (AW217982, AW041695, AW030725, BF096782), HsfA5b (AW034402), HsfA6b (AW036683, AW932142, AW222011, BE 434585, BE433803, AI895294, AI489721, BG132247), HsfA6c (BG351853), HsfA8a (AW738023, mosaic Hsf, whose annotation to the HsfA8 group depends on the C-terminal part of the DBD and the HR-A/B region), HsfA8b (AW931892, not included, because only C-terminal part with HR-A/B region available), HsfB1 (BF097217, BG134658, AI895934), HsfB2a (AW931781, AW220758, AW931176), HsfC1 (AW738534, AW979619, BE451302, AW649243); G max HsfA2a (Hsf21 fragment, Z46952), HsfA2b (AW164509), HsfA4 (BE611683, AW756148, BG405291, BE330669), HsfA6 (BG041837), HsfB2a (BE346810, BF067962), HsfB2b (AW703969), HsfB3 (BE019974), HsfB4b (BF597135), HsfC1 (BE347442, AW596493, BG352891); S tuberosum HsfA5 (BF459947), HsfB2a (BE473183), HsfB2b (BF052865), HsfC1 (BF459641); H vulgare HsfA2a (BE603513), HsfA4 (BE216310), HsfA2b (BF264338), HsfC1 (BF616419); Zea mays: ZmHsfA2/A8 (Hsfa fragment, S61458)