Abstract
The only animal cells known that can maintain functional plastids (kleptoplasts) in their cytosol occur in the digestive gland epithelia of sacoglossan slugs. Only a few species of the many hundred known can profit from kleptoplasty during starvation long-term, but why is not understood. The two sister taxa Elysia cornigera and Elysia timida sequester plastids from the same algal species, but with a very different outcome: while E. cornigera usually dies within the first two weeks when deprived of food, E. timida can survive for many months to come. Here we compare the responses of the two slugs to starvation, blocked photosynthesis and light stress. The two species respond differently, but in both starvation is the main denominator that alters global gene expression profiles. The kleptoplasts' ability to fix CO2 decreases at a similar rate in both slugs during starvation, but only E. cornigera individuals die in the presence of functional kleptoplasts, concomitant with the accumulation of reactive oxygen species (ROS) in the digestive tract. We show that profiting from the acquisition of robust plastids, and key to E. timida's longer survival, is determined by an increased starvation tolerance that keeps ROS levels at bay.
Keywords: invertebrates, sacoglossa, kleptoplasty, reactive oxygen species, starvation, photosynthetic slugs
1. Introduction
Some sacoglossan slugs can house functional plastids for months in the cytosol of cells that line the animals' digestive tubules. The slugs steal the plastids (kleptoplasts) from siphonaceous algae upon which they feed. Theory has it that the presence of functional kleptoplasts [1,2] allows some of the sea slug species to survive starvation periods that can last almost a year [3,4]. Recent reports question the categorical importance of photosynthesis during starvation for several species [5,6] and while the animals fix CO2 in a light-dependent manner [5,7–10], for how long and how much during starvation is not well documented. Animals induce autophagy to reallocate resources through the recycling of tissue when facing starvation [11] and mitochondria-generated reactive oxygen species (ROS) signalling has been found to be a key mediator of autophagy triggered by nutrient deprivation [12,13]. Over the past 20 years, research on ‘photosynthetic (green) slugs’ has focused on understanding how the stolen plastids can remain functional in a foreign cytosol [14], but the molecular response to starvation has not, to our knowledge, been assessed in sacoglossan slugs until now.
The kleptoplasts' stability outside of the algae disagrees with our knowledge about the sensitivity of land plant plastids, whose photosystem II is highly susceptible to photosynthesis-associated damage [2,15,16]. Recent works [16,17] are reviving old observations that demonstrated some algal plastids are naturally more robust than land plant plastids [18]. Kleptoplast longevity therefore appears an intrinsic property the stolen organelles bring along, and depending on the slug species this results in different modes of kleptoplast retention and lengths of survival during starvation [2,16]. It is generally suspicious that among 300 sacoglossan species that feed on siphonaceous algae, only seven species are known to maintain functional kleptoplasts and survive long term [2,19,20].
The two congeneric species Elysia cornigera and Elysia timida both feed on the ulvophycean alga Acetabularia, but only E. timida tolerates prolonged starvation [2,21]. Why? We compared the two sister taxa by monitoring their kleptoplasts' photosynthetic capacity and characterizing the slugs' physiological response to starvation focusing on gene expression modulation and ROS development. Our results indicate that algal cytosol are the slug's main food source and furthermore demonstrate that the plastid-bearing slugs' capacity to endure prolonged periods of starvation is not determined by the photosynthetic activity of their kleptoplasts, but by how they have evolved to respond to starvation, in particular the ability to cope with ROS.
2. Results and discussion
(a). Performance of stolen plastids does not depend on the slug species
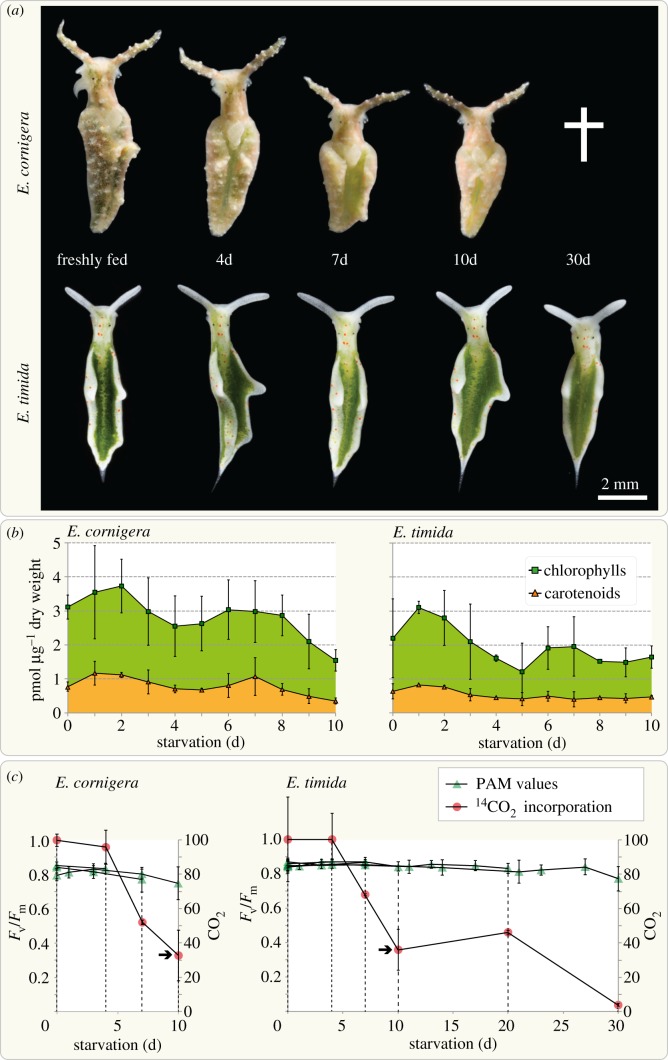
We fed E. timida and E. cornigera solely on Acetabularia acetabulum DI1 from the day they hatched in our laboratory. After eight and six weeks, respectively, and at which time the animals had reached maturity, the slugs were separated from the alga and routinely kept at 25 µmol quanta m−2 s−1 for 12 h every day. All sacoglossans that starve are observed to shrink [10,22] and the same was true for our two species. Yet, E. cornigera decreased in size more rapidly than E. timida (figure 1a) and rarely survived more than two weeks without food, which is consistent with previous reports [19,21]. Loss in size was accompanied by a slow fading of the animals' greenish coloration owing to the loss of the pigments chlorophyll (Chl) b and, most prominently, Chl a (electronic supplementary material, table S1). In E. cornigera, pigment concentration declined from starvation day 2 to 4 by 32.9%, but then increased again for a few days in relation to the dry weight of the animals (figure 1b). At day 10, the Chl a and b concentration had dropped from 3109 to 1544 pmol mg−1 animal dry weight (49.7%). These values correlate with the rapid decrease observed in body size between day 4 and 7 of starvation (figure 1a). In E. timida, we observed a similar, yet less steep, decline in the concentration of pigments in relation to body mass over the 10 days of starvation measured (from 2193 to 1606 at day 4 (73.2%) and 1638 pmol Chl a + b/mg animal dry weight at day 10 (74.7%); figure 1b). The increase of photosynthesis-associated pigments around day 6 demonstrates that the animals we measured metabolize their own tissue at an apparently faster rate than the kleptoplasts degrade, which was also observed for starving juveniles of E. chlorotica [23].
Figure 1.
Elysia cornigera dies while showing comparable photosynthetic activity to E. timida during starvation. Adult slugs were fed on Acetabularia acetabulum and the effects of starvation at 25 µmol quanta m−2 s−1 on the photosynthetic capacity of both species evaluated. (a) Representative images of one single specimen of each species. Elysia cornigera shrinks more rapidly than E. timida, and while the E. cornigera specimens never survived starvation for more than two weeks, all E. timida specimens survived and retained an almost equal body length during the 30 d of starvation analysed. (b) Analysis of pigment concentration in relation to dry weight of the slugs during the first 10 days of starvation. Quantification was performed on three biological replicates per time point and normalized to an external standard; each pigment extract was derived from three to seven E. cornigera (in total 143 individuals) and two E. timida individuals (in total 66 individuals). (c) Assessment of the maximum quantum yield (Fv/Fm; measured using a PAM fluorometer) of photosystem II and net 14CO2 fixation relative to freshly fed animals (t0), during starvation within the same group of individuals. Each single Fv/Fm curve represents measurements on a group of at least 12 E. cornigera and nine E. timida specimens; from each group four E. cornigera (in total 48 individuals) and three E. timida (in total 54 individuals) were used for determining the 14CO2 incorporation after 0, 4, 7, 10, 20 and 30 days of starvation (latter two only for E. timida).
Photosynthetic capacity was determined through measurements of photosystem II quantum efficiency (derived from Chl fluorescence analyses determined by pulse-amplitude modulation (PAM) fluorometry) and the amount of incorporated 14CO2. Freshly fed slugs showed a similar initial maximum quantum yield (Fv/Fm of 0.83 ± 0.03 for E. cornigera and 0.85 ± 0.03 for E. timida). Measured Fv/Fm of E. timida declined slowly (−0.001 Fv/Fm per day starved) over the one-month period, reaching 0.77 ± 0.07 at day 30. In E. cornigera, the decline of Fv/Fm was more prominent (−0.008 Fv/Fm per day starved) over the 10 days measured, dropping to 0.75 ± 0.1, at which point the species began to die (figure 1c). At the same time, the amount of 14CO2 incorporation had dropped to 34% (±15%) and 36% (±12%) in E. cornigera and E. timida, respectively, after 10 days of starvation (figure 1c). At that point, the Fv/Fm had dropped to 0.84 ± 0.03 and 0.75 ± 0.1 for E. timida and E. cornigera, respectively. After 30 days of starvation, the amount of 14CO2 fixed in E. timida had dropped to 4 ± 1% in regard to the initial amount. In any case, after 10 days of food deprivation E. cornigera died in the presence of intact (electronic supplementary material, figure S1) and functional kleptoplast. The difference of profiting from kleptoplasty must depend on the animals' tolerance to starvation, not the stolen organelles' performance. This observation challenges the idea that kleptoplasts could act as food depot [5,10], and raised the question, why in particular E. cornigera dies in the presence of functional kleptoplasts.
(b). Global gene expression response is predominantly governed by starvation
To uncover the differences of the two species in the response to starvation, and test whether the slugs can to some respect sense the kleptoplasts' status, we analysed gene expression changes throughout starvation and under different environmental stimuli. We performed comparative transcriptomics of the slugs: (i) under starvation (S), (ii) under starvation and in the presence of the photosynthesis inhibitor drug monolinuron (S + M), and (iii) under starvation and including a daily bleaching pulse with 1000 µmol quanta m−2 s−1 for 1 h d−1 (S + B). A total of 10 857 contigs for E. cornigera and 11 152 contigs for E. timida were assembled that were supported by at least 100 reads and homologous to eumetazoan sequences (electronic supplementary material, figure S2 and table S2). Global gene expression trends were confirmed for all conditions on six individual genes using quantitative reverse-transcription PCR (qRT-PCR) (electronic supplementary material, figure S3).
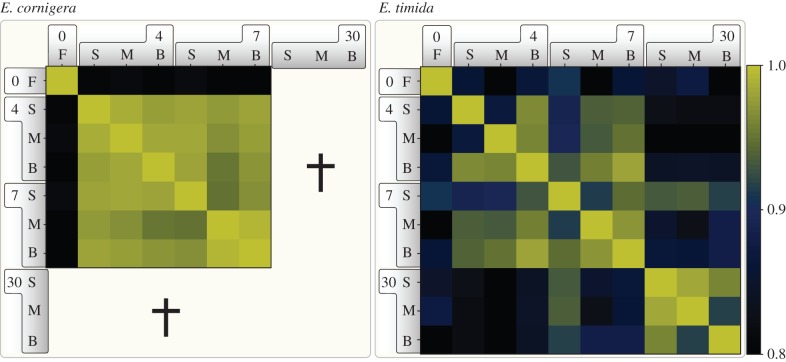
If the slugs have evolved to depend on the photosynthetic capacity of their kleptoplasts, we would expect to see a strong difference in the regulation of global gene expression profiles when kleptoplast electron transport is blocked (S + M) or when the kleptoplasts are under high light stress (S + B). Yet, when analysing the co-regulation of gene expression we found that starvation (S) in comparison to feeding (F) was the main denominator for the transcription response in E. cornigera (figure 2). The additionally applied treatments (S + B, S + M) only had marginal effects on the global expression profile of E. cornigera compared to starvation alone (S). In contrast to this, the response of E. timida throughout the first two weeks of starvation is more differentiated in regard to the individual stimuli and in comparison to E. cornigera (figure 2). After 4 d of S + M and 7 d S + B, the most differing expression was detected, indicating that E. timida indeed responded to the status of their kleptoplasts. Intriguingly, for E. timida a similar yet delayed transcriptional response to starvation was observed, as transcriptional profiles at day 30 began to reflect those observed for E. cornigera earlier during starvation based on the correlation of co-regulated genes within each species (figure 2). The slugs predominantly respond to starvation and not to the photosynthetic capacity of the kleptoplasts.
Figure 2.
Global co-expression analysis of gene expression changes. Pearson correlations were calculated by comparing the FPKM (fragments per kilobase of exon per million reads mapped) values of filtered genes of the different samples. Note the clear separation of the control and the remaining conditions and for E. timida the shift between the time points of 4 and 7 days in comparison to 30 days. The colour key indicates the degree of co-regulation. The bold numbers represent time of treatment in days. No values for E. cornigera were obtained for 30 days, because all individuals had succumbed to starvation (indicated by a cross). F, freshly fed; S, starving; M, monolinuron; B, bleach.
(c). Host response underpins kleptoplast compatibility
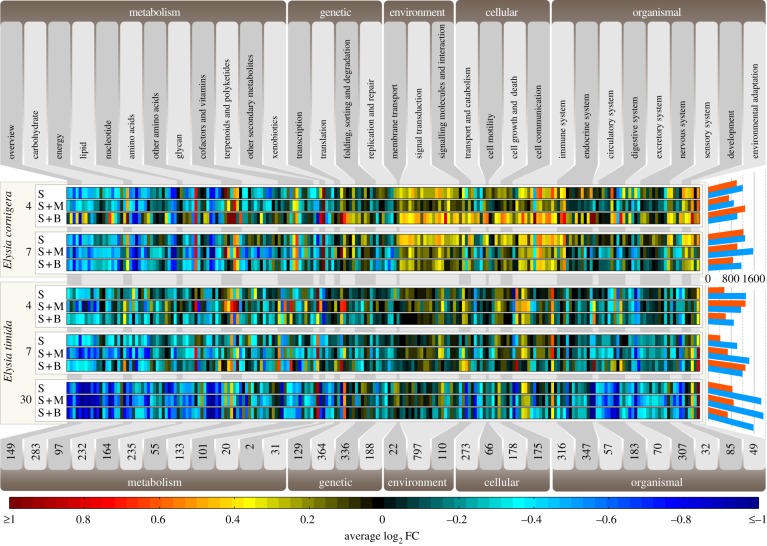
When mapping the transcriptomes to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (figure 3), two main trends were observed: first, downregulation of carbohydrate, energy, and lipid metabolism associated pathways (mean log2FC of −0.32 versus an overall mean log2FC of −0.10). On average, 81 of the 89 metabolism pathways detected for E. timida were downregulated after 30 days of starvation. Monolinuron, blocking the quinon-binding site of the D1 protein [24], enforced the downregulation of core metabolic genes in both species (at 7 d S + M log2FC −0.44 in both). Second, in contrast to E. timida (mean log2FC of −0.11), starvation in E. cornigera triggered an elevated expression of environmental information processes and cellular and organismal systems including stress response and autophagy (mean log2FC 0.13). Light stress further promoted this reaction in E. cornigera (mean log2FC of 0.20 at 4 d/7 d S + B) compared to E. timida (mean log2FC of –0.03 at 4d/7d S + B). Starvation imposes a metabolic dormancy and gene expression profiles to a minor degree reflect kleptoplast manipulation. The activity of the kleptoplasts, however, does not explain the survival during starvation.
Figure 3.
Gene expression changes of animal KEGG pathways. Transcriptomes were sequenced for both animals after 4 and 7 days of starvation (bold numbers), and additionally after 30 days for E. timida. Elysia cornigera and E. timida were subjected to three different conditions: starvation alone (S), starvation plus monolinuron (S + M; 2 µg ml−1) and starvation plus bleaching (S + B; 1000 µmol quanta m−2 s−1 for 1 h each day). Upregulation is shown from red to black and downregulation from black to blue. The numbers at the bottom represents the amount of K-numbers (KEGG orthology group identifier) found within each reaction module (modules in alternating light and grey flags). The bars on the right show the total number of regulated genes (minimum of twofold) for the individual conditions analysed (blue bars, downregulation; red bars, upregulation).
Ubiquitination is a key mechanism to tag organelles and misfolded proteins—accumulating under stress conditions such as starvation—for degradation [25,26]. Expression levels of polyubiquitin remained the same in both species, except for a single polyubiquitin homolog of E. timida (Eti000028, 4.5-fold upregulation, p < 0.05). For the ubiquitin binding autophagic receptor p62/SQSTM1 [27], an average of only 28 FPKMs (fragments per kilobase of exon per million reads mapped) were identified for E. cornigera, but 659 FPKMs for E. timida. And only for the latter species an upregulation of p62/SQSTM1 was observed during starvation (electronic supplementary material, table S3). Elysia timida might be better equipped to detect damaged proteins and organelles than E. cornigera, allowing the former to better recognize and remove damaged organelles and misfolded hazardous proteins during starvation. The ability to recognize damaged kleptoplasts for their subsequent degradation could explain how the slugs profit from stored kleptoplasts. Only malfunctioning and ROS-leaking plastids would be detected by the autophagosomal machinery and removed; the profit of kleptoplasts would be an indirect one. This would also explain the more rapid decline of Fv/Fm values in E. cornigera in comparison to E. timida, while retaining the same CO2 fixation rate (figure 1c). Accumulation of damaged plastids that fix no CO2 would lead to a lower Fv/Fm ratio in E. cornigera. If E. timida specifically removes damaged plastids, then all malfunctioning photosystems are excluded from that equation.
We found that the upregulation of genes encoding ROS scavengers differed between the two sister taxa. With progressing starvation, E. cornigera elevated the expression of superoxide-dismutase homologs regardless of the treatment (e.g. Eco000152; on average 3.1-fold up at day 7, p < 0.05). Apparent changes in the expression of catalases were only detected for E. timida in the first days of starvation (Eti001444; up more than twofold on average). It was recently noted that a crucial part of the slug-plastid symbiosis is evolving mechanisms to cope with kleptoplast-associated toxins [2]. A difference in stress response and ROS evolution could explain the different survival rates of the two species observed during starvation.
(d). Elysia cornigera accumulates reactive oxygen species, not Elysia timida
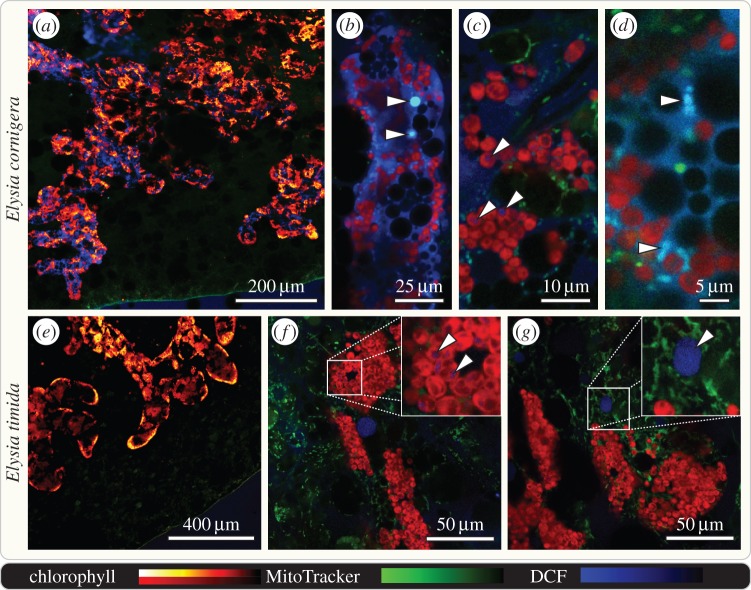
ROS are known as signalling molecules [13] but also as deleterious cytotoxins [28]. To monitor hydrogen peroxide (H2O2) production on tissue level we used 2′,7′-dichlorofluorescin (DCF). Freshly fed slugs showed no detectable amount of H2O2 (electronic supplementary material, figure S4), but at day 10 of starvation H2O2 was most prominently detected in E. cornigera within the lumen of the digestive tubules (figure 4a,e). Both the lumen and the epithelium of the digestive tubules—the localization of the kleptoplasts—showed intensive DCF staining (figure 5). By contrast, DCF staining in E. timida occurred only in some globular structures of 10–20 µm in diameter, but of otherwise unknown nature, within individual digestive tubules (figure 4g, arrowhead). After 30 d of starvation, the digestive glandular tubules of E. timida still showed no apparent increase in H2O2 levels (electronic supplementary material, figure S4), but DCF now additionally stained globular structures of approximately 100 µm in diameter that were situated between the epidermis and the digestive tubules, and independent from the latter. These structures could represent one of the secondary metabolite-harbouring vacuoles typical for these kind of slugs [29]. Stroma of the A. acetabulum kleptoplasts resting within the digestives tubule cells of both E. cornigera and E. timida showed equal intensity in regard to H2O2 levels (figure 4c, f insert). Mitochondria, a common source of ROS [13], were found to accumulate H2O2 as proved through the co-localization of the DCF signal with MitoTracker Red (figure 4d). Hydrogen peroxide, a critical ROS in terms of cytotoxicity [28], accumulates only in E. cornigera.
Figure 4.
Hydrogen peroxide accumulates in the digestive tubules of starving E. cornigera, but not E. timida. Confocal laser scanning micrographs of the kleptoplast-bearing digestive tubules that pervade the parapodia of 10 days starved, whole mount E. cornigera and E. timida stained with 100 µM dichlorofluorescin (DCF; blue) and MitoTracker Red (green); chlorophyll autofluorescence in red to yellow (a,e) or red only (b–d, f, g). Digestive tubules of E. cornigera (lined with autofluorescent kleptoplasts) show well-defined DCF staining within the lumen and epithelium (a–d). Details of single digestive tubules revealing readily H2O2 accumulators of equal size as kleptoplasts (b arrowheads), kleptoplasts displaying weak DCF staining (c; see arrowheads) and MitoTracker Red co-localizing with DCF staining (d). Digestive tubules of E. timida displaying no distinguishable DCF signal in epithelium or lumen (e). Details of single digestive tubules revealing accumulating weak levels of H2O2 (f, arrowheads), comparable to those seen in E. cornigera (c, arrowheads) and showing globular structures of unknown nature accumulating H2O2 (g, arrowhead).
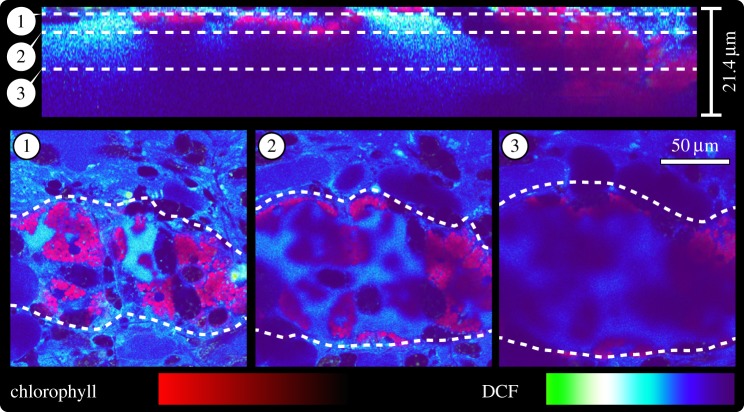
Figure 5.
Hydrogen peroxide is present in the lumen and epithelium of digestive tubules of starving E. cornigera. Orthogonal view of a confocal laser scanning z-stack micrograph reconstructed from 58 individual optical sections (21.35 µm of depth in total) of a single digestive tubule (outlined by dashed lines in the bottom panel) of 10d starved E. cornigera stained with 100 µM dichlorofluorescin (DCF; purple-blue to green gradient); chlorophyll autofluorescence in red. At the top, a transverse section trough the stack is shown to illustrate at which level the three coronal sections (indicated by dashed lines) were captured.
Animals respond to nutrient depletion by autophagy, and superoxide (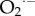 ) is a key signal that triggers the autophagosomal machinery [12,13] and mammalian tissues cultures show strong increase in ROS after a few hours of starvation [12]. We conducted nine separate experiments, each time with different individuals, and monitored
) is a key signal that triggers the autophagosomal machinery [12,13] and mammalian tissues cultures show strong increase in ROS after a few hours of starvation [12]. We conducted nine separate experiments, each time with different individuals, and monitored 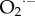 evolution using dihydroethidium (DHE) which stains the nuclei of cells that experience elevated levels of
evolution using dihydroethidium (DHE) which stains the nuclei of cells that experience elevated levels of 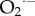 [30]. Chl autofluorescence was observed to decrease, but not completely fade, in both species throughout the 10 d monitored (figure 6a). This concurs with the pigment measurements and the ability of the remaining kleptoplasts to incorporate 14CO2 (figure 1c). Feeding slugs showed only faint
[30]. Chl autofluorescence was observed to decrease, but not completely fade, in both species throughout the 10 d monitored (figure 6a). This concurs with the pigment measurements and the ability of the remaining kleptoplasts to incorporate 14CO2 (figure 1c). Feeding slugs showed only faint 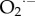 signals towards the edges of the parapodia, especially the epidermis (figure 6a). Staining did not predominantly dye nuclei of the plastid-bearing digestive gland cells, but instead those of surrounding and connective tissue of the parapodia. Levels of
signals towards the edges of the parapodia, especially the epidermis (figure 6a). Staining did not predominantly dye nuclei of the plastid-bearing digestive gland cells, but instead those of surrounding and connective tissue of the parapodia. Levels of 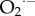 , apparent through the number of nuclei stained and the intensity of the staining itself, steadily increased throughout starvation and predominantly in E. cornigera (figure 6a). While both slug species harboured functional kleptoplasts stemming from the same source, only E. cornigera exhibited increasing levels of
, apparent through the number of nuclei stained and the intensity of the staining itself, steadily increased throughout starvation and predominantly in E. cornigera (figure 6a). While both slug species harboured functional kleptoplasts stemming from the same source, only E. cornigera exhibited increasing levels of 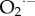 that were significant, but not E. timida (p < 0.001). Elysia cornigera accumulates ROS to a much higher degree than its sister taxon (figure 6b), reflecting the different capacities of the two species with regard to managing starvation in the presence of functional kleptoplasts.
that were significant, but not E. timida (p < 0.001). Elysia cornigera accumulates ROS to a much higher degree than its sister taxon (figure 6b), reflecting the different capacities of the two species with regard to managing starvation in the presence of functional kleptoplasts.
Figure 6.
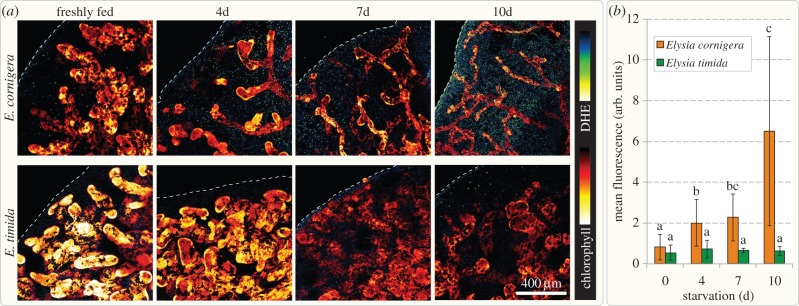
Superoxide accumulation in starving slugs. (a) Confocal laser scanning micrographs of the kleptoplast-bearing digestive tubules from similar positions on the parapodia of freshly fed and 4, 7 and 10 days starved, whole mount E. cornigera and E. timida (optical sections; pinhole set to 1 airy unit) stained with 100 µM DHE (staining 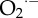 ), shown in blue-yellow gradient; chlorophyll autofluorescence of the kleptoplasts is shown as red to yellow glow. Dashed lines mark the outer rim of the parapodia. DHE signal increases over time in E. cornigera, while the diameter of the digestive tubules decreases, visible due to their lining with kleptoplasts. Superoxide in the parapodia of E. timida does not significantly increase over time, while kleptoplasts lining the digestive tubules appear less densely packed. (b) Quantification of the mean DHE fluorescence intensity in the parapodial tissue; significance groups (for (a,b) p < 0.01; (c) p < 0.05) were determined using a Mann–Whitney U-test; measurement of fluorescence intensity was performed on greater than or equal to six individual specimens for each time point; error bars indicate standard deviation.
), shown in blue-yellow gradient; chlorophyll autofluorescence of the kleptoplasts is shown as red to yellow glow. Dashed lines mark the outer rim of the parapodia. DHE signal increases over time in E. cornigera, while the diameter of the digestive tubules decreases, visible due to their lining with kleptoplasts. Superoxide in the parapodia of E. timida does not significantly increase over time, while kleptoplasts lining the digestive tubules appear less densely packed. (b) Quantification of the mean DHE fluorescence intensity in the parapodial tissue; significance groups (for (a,b) p < 0.01; (c) p < 0.05) were determined using a Mann–Whitney U-test; measurement of fluorescence intensity was performed on greater than or equal to six individual specimens for each time point; error bars indicate standard deviation.
3. Conclusion
Contrary to the current view our study demonstrates that the presence of functional kleptoplasts alone does not underpin the survival of green photosynthetic slugs. It is first and foremost the ability of some species to tolerate starvation. The best hints we currently have that underpin starvation tolerance include the upregulation of a set of canonical ROS quenchers such as catalases, and/or slug-specific polyproprionate metabolites such as Elysiapyrone or Elysione [31], and/or the efficient and specific degradation of damaged kleptoplasts. Literature has divided slugs into non-, short- and long-term retention species, depending on for how long kleptoplasts are functionally retained [19]. Our results demonstrate that it is equally important to divide the slugs into starvation-intolerant and starvation-tolerant species. That tolerance might be associated with the habitat and availability of the food source. Elysia cornigera was collected from the Caribbean Sea, but E. timida from the Mediterranean. Maybe only E. timida experiences food shortage due to seasonal differences and a difference in the calcification of individual algal species, and had to evolve accordingly. In any case, sacoglossan research has focused on how the animals chaperone their kleptoplasts, but it should just as much focus on how they manage starvation, because it will teach us on how they suppress ROS stress.
4. Experimental procedures
Elysia timida was collected on Giglio, Italy (42°22′ N, 10°52′ E and 42°21′ N, 10°52′ E) between 3 and 6 m depth. Elysia cornigera was collected at the Florida Keys (24°38′ N, 81°18′ W) at about 1 m depth. Both species were kept at a 12 L : 12 D rhythm at 25 µmol quanta m−2 s−1 in 3.7% salt water (Tropic Marine) and at 21°C. Acetabularia acetabulum DI1 (isolate of Professor Diedrik Menzel, Bonn University, Germany) was grown at the same conditions as the animals, but in algal f/2 medium. For starvation experiments, animals were moved to Petri dishes (∅︀200 mm) and 50% of the water was changed every other day. Monolinuron (JBL GmbH) was added from a stock solution of 4000 mg l−1 to a final concentration of 2 µg ml−1. Bleaching was performed by illuminating the slugs with 1000 µmol quanta m−2 s−1 for 1 h every day at the same time. For each pigment extraction, pooled slugs were transferred to 90% acetone for each time point, homogenized and kept at −20°C for 1 day. Extractions for every time point were carried out as biological triplicates, each replicate consisting of at least three E. cornigera and two E. timida. Crude extracts were filtered through a 200 nM polytetrafluoroethylene membrane and then analysed by reversed-phases high pressure liquid chromatography with ultraviolet/visible spectroscopy detection (Hitachi/Merk) as described earlier [32]. Pigment concentrations were determined using external pigment standards isolated from spinach thylakoids. Dry weight was determined on dried slug homogenate after extraction.
Photosystem II maximum quantum yield (Fv/Fm) was determined using a WALZ MINI-PAM as described previously [33], on the same group of individuals that were used for 14CO2 incorporation measurements. For every time point for which 14CO2 incorporation was determined, an individual group of slugs (at least 12 E. cornigera or nine E. timida) was kept in isolated dishes. For each of these groups, separate Fv/Fm measurements were performed, and each individual slug of a group was measured at least three times. For each data point, the mean of all individual triplicate measurements was calculated. The light-driven incorporation of 14CO2 was determined as described earlier [5]. Briefly, for each triplicate measurement four individuals of E. cornigera and three of E. timida were used. Slugs were incubated in 1.2 ml artificial seawater supplemented with 0.40 mM [14C]-NaHCO2 (25 µCi per incubation, NEN-radiochemicals, MA, USA) for 2 h at room temperature and 72 µmol quanta m−2 s−1 illumination. After rinsing, homogenization and acidification of the material with 150 µl 1 M HCl, all the substrate was expelled from the homogenate. Incorporation of 14C by the slugs was determined in a scintillation counter after the addition of 12 ml LUMA-Gel scintillation cocktail (LUMAC, The Netherlands).
For ROS-imaging, slugs were incubated for 45 min with 100 µM DHE (DHE; excitation/emission HeNe 543/610 nm; Sigma) or 100 µM DCF (2′,7′-dichlorofluorescin; excitation/emission Argon 488/529 nm) plus 2 µM MitoTracker Red CMXRos (excitation/emission HeNe 543/599 nm; LifeTechnologies) in artificial seawater. Slugs were rinsed twice with seawater and decapitated before mounting. Confocal laser scanning microscopy was carried out using a Zeiss LSM 710. Imaging was always performed with the same settings and at a similar position along the parapodial rim at the same depth relative to the epidermis. Images were processed using Fiji/ImageJ 1.48f [34] and statistics were performed using R [35]. Normality was tested via a Shapiro–Wilk test [36] and significance using a Mann–Whitney U-test [37].
Total RNA was extracted three times from seven E. cornigera (a total of 21) and three times from five E. timida (a total of 15) feeding on A. acetabulum (t0, feeding control group), 28 individuals of E. cornigera that were starved for 4 days, of which nine individuals had only starved (S), nine had starved and were treated with monolinuron (S + M), and 10 of which had starved and were treated with 1000 µmol quanta m−2 s−1 for 1 h each day (S + B). The RNA of 20 E. timida's (7 S, 7 S + M, 6 S + B) was isolated after four days of starvation (t1), of 30 E. cornigera (10 S, 9 S + M, 11 S + B) and of 20 E. timida (6 S, 8 S + M, 6 S + B) after 7 days of starvation (t2), and finally of 32 E. timida (11 S, 10 S + M, 11 S + B) after 30 days of starvation (t3). RNA was isolated using TRIzol (Life Technologies) according to the manufacturer's protocol and an additional DNAse treatment (Thermo Fisher Scientific). Poly(A) mRNA enrichment, library preparation using the TruSeq kit (Illumina) and 100 bp paired end sequencing using the Illumina HiSeq2000 system was performed by the Beijing Genome Institute (Hong Kong). Reads with remaining adapter sequences, more than 5% of unknown nucleotides or more than 20% of nucleotides with quality scores less than 10 (Illumina 1.5) were removed as part of the raw data extraction.
A total of 1 186 405 486 reads (a minimum of 52 million reads/library) were obtained (electronic supplementary material, table S2). Reads were inspected by FastQC v. 0.10.1 [38] and filtered and trimmed using Trimmomatic 0.32 [39] (parameters: ILLUMINACLIP:TruSeq3-PE.fa:2 : 30 : 10; HEADCROP:10; TRAILING:3; SLIDINGWINDOW:4 : 20; MINLEN:36). Reads of all samples were assembled using Trinity r20131110 [40], which yielded 249 855 and 150 314 transcripts for E. cornigera and E. timida, respectively. Expression values for transcript clusters (or unigenes) were extracted and analysed using the Trinity pipeline [41] via RSEM 1.2.11 [42], Bowtie 1.0.1 [43] and edgeR 3.4.2 [44]. Only unigenes with raw read counts of greater than or equal to 100 over at least two samples were analysed, resulting in 36 380 and 32 897 unigenes, respectively. Differential expression was determined by log2fc of at least ±1 compared to the control set of unigenes (with raw read counts of greater than or equal to 100). Expression change significance was assessed based upon the dispersion of available replicate information in edgeR. The longest isoform of each cluster was defined as the representative sequence for a unigene.
To plot the overall taxonomic distribution of the assembled transcripts, unigenes were subjected to a BLASTx-based [45] search (e-value cut-off 10−10) against a database consisting of protein sequences of RefSeq version 64 [46] plus those of the genomes of Crassostrea gigas [47] and Lottia gigantean [48] (electronic supplementary material, figure S2). For all downstream analyses, only those genes were included for which top hits to the mentioned organisms or other metazoans were retrieved. Excluded were those with best blast hits to plants, protists, prokaryotes and viruses. Protein annotations for the 14 848 E. cornigera and 13 875 E. timida unigenes were extracted from mentioned BLAST hits and from a second BLASTx search to the UniProtKB/Swiss-Prot database [49]. KOG/NOG (EuKaryotic/Non-supervised Orthologous Groups [50–52]) categories were determined based on the best BLASTx hit to protein sequences of eggNOG v. 4 [53]. KEGG [54] accessions were obtained using the KAAS 1.6a webservice [55] against metazoa.
For qRT-PCR, 200 ng RNA was reverse transcribed using random hexamers and the iScript Select cDNA Synthesis Kit (Bio-Rad). Two-step qRT-PCR was performed on biological triplicates for each time point and treatment (each containing pooled RNA from greater than or equal to five slugs) using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer's instructions on a StepOne Plus (Applied Biosystems) real-time PCR system. Primers were designed using Primer-BLAST [56] (for primer sequences, see the electronic supplementary material, table S4) and data were analysed according to Pfaffl [57], using the EcRPL38 (Eco000149), EtRPL19 (Eti000121) and EtSETMAR (Eti000317) as reference genes.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Margarete Stracke for assistance in culturing, Steffen Köhler (CAi, HHU) for help with slug photography and Sophie Rommel (Population Genetics, HHU) for help with the statistical analysis. We thank the ‘Zentrum für Informations- und Medientechnologie’ (ZIM, HHU Düsseldorf) for computational support and infrastructure.
Data accessibility
All sequencing data was deposited at the NCBI Single Read Archive (SRP047359) and the Transcriptome Shotgun Assembly Sequence Database (E. cornigera: GBRW00000000; E. timida: GBRM00000000).
Author contributions
J.d.V., A.G.M.T., P.J. and S.B.G. carried out the main experiments. G.C., J.d.V. and H.W. additionally collected animals and C.W. processed the RNA-seq data. S.B.G. designed the experiments and wrote the manuscript together with J.d.V. All authors discussed the results, read and approved the final manuscript.
Funding statement
Funding through the Deutsche Forschungsgemeinschaft to S.B.G. (GO1825/4-1) is gratefully acknowledged.
References
- 1.Trench RK. 1969. Chloroplasts as functional endosymbionts in the mollusc Tridachia crispate (Bërgh), (Opisthobranchia, Sacoglossa). Nature 222, 1071–1072. ( 10.1038/2221071a0) [DOI] [Google Scholar]
- 2.de Vries J, Christa G, Gould SB. 2014. Plastid survival in the cytosol of animal cells. Trends Plant Sci. 19, 347–350. ( 10.1016/j.tplants.2014.03.010) [DOI] [PubMed] [Google Scholar]
- 3.Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. 2008. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl Acad. Sci. USA 105, 17 867–17 871. ( 10.1073/pnas.0804968105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz S, Calado R, Serôdio J, Cartaxana P. 2013. Crawling leaves: photosynthesis in sacoglossan sea slugs. J. Exp. Bot. 64, 3999–4009. ( 10.1093/jxb/ert197) [DOI] [PubMed] [Google Scholar]
- 5.Christa G, Zimorski V, Woehle C, Tielens AGM, Wägele H, Martin WF, Gould SB. 2014. Plastid-bearing sea slugs fix CO2 in the light but do not require photosynthesis to survive. Proc. R. Soc. B 281, 20132493 ( 10.1098/rspb.2013.2493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klochkova TA, Han JW, Chah K-H, Kim RW, Kim J-H, Kim YK, Kim GH. 2013. Morphology, molecular phylogeny and photosynthetic activity of the sacoglossan mollusc, Elysia nigrocapitata, from Korea. Mar. Biol. 160, 155–168. ( 10.1007/s00227-012-2074-7) [DOI] [Google Scholar]
- 7.Trench RK, Boyle JE, Smith DC. 1973. The association between chloroplasts of Codium fragile and the mollusc Elysia viridis. Proc. R. Soc. Lond. B 184, 63–81. ( 10.1098/rspb.1973.0031) [DOI] [Google Scholar]
- 8.Clark KB, Jensen KR, Stirts HM, Fermin C. 1981. Chloroplast symbiosis in a non-elysiid mollusc Costasiella lilianae Marcus (Hermaeidae: Ascoglossa (=Sacoglossa): effects of temperature, light intensity, and starvation on carbon fixation rate. Biol. Bull. 160, 43–54. ( 10.2307/1540899) [DOI] [Google Scholar]
- 9.Marín A, Ros M. 1989. The chloroplast-animal association in four Iberian sacoglossan Opisthobranchs: Elysia timida, Elysia translucens, Thuridilla hopei and Bosellia mimetica . Scient. Mar. 53, 429–440. [Google Scholar]
- 10.Christa G, de Vries J, Jahns P, Gould SB. 2014. Switching off photosynthesis: the dark side of sacoglossan slugs. Commun. Integr. Biol. 7, e28029 ( 10.4161/cib.28029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh R, Cuervo AM. 2011. Autophagy in the cellular energetic balance. Cell. Metab. 13, 495–504. ( 10.1016/j.cmet.2011.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760. ( 10.1038/sj.emboj.7601623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Chen Y, Gibson SB. 2013. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell. Signal. 25, 50–65. ( 10.1016/j.cellsig.2012.09.020) [DOI] [PubMed] [Google Scholar]
- 14.Pelletreau KN, Bhattacharya D, Price DC, Worful JM, Moustafa A, Rumpho ME. 2011. Sea slug kleptoplasty and plastid maintenance in a metazoan. Plant Phys. 155, 1561–1565. ( 10.1104/pp.111.174078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickelsen J, Rengstl B. 2013. Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635. ( 10.1146/annurev-arplant-050312-120124) [DOI] [PubMed] [Google Scholar]
- 16.de Vries J, Habicht J, Woehle C, Huang C, Christa G, Wägele H, Nickelsen J, Martin WF, Gould SB. 2013. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol. Evol. 5, 2540–2548. ( 10.1093/gbe/evt205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jesus B, Ventura P, Calado G. 2010. Behaviour and a functional xanthophyll cycle enhance photo-regulation mechanisms in the solar-powered sea slug Elysia timida (Risso, 1818). J. Exp. Mar. Biol. Ecol. 395, 98–105. ( 10.1016/j.jembe.2010.08.021) [DOI] [Google Scholar]
- 18.Giles KL, Sarafis V. 1972. Chloroplast survival and division in vitro. Nat. New Biol. 236, 56–58. ( 10.1038/newbio236056a0) [DOI] [PubMed] [Google Scholar]
- 19.Händeler K, Grzymbowski YP, Krug PJ, Wägele H. 2009. Functional chloroplasts in metazoan cells: a unique evolutionary strategy in animal life. Front. Zool. 6, 28 ( 10.1186/1742-9994-6-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christa G, Gould SB, Franken J, Vleugels M, Karmeinski D, Händeler K, Martin WF, Wägele H. 2014. Functional kleptoplasty in a limapontioidean genus: phylogeny, food preferences and photosynthesis in Costasiella with a focus on C. ocellifera (Gastropoda: Sacoglossa). J. Mollusc. Stud. 80, 499–507. ( 10.1093/mollus/eyu026) [DOI] [Google Scholar]
- 21.Krug PJ, Händeler K, Vendetti J. 2011. Genes, morphology, development and photosynthetic ability support the resurrection of Elysia cornigera (Heterobranchia: Plakobranchoidea) as distinct from the ‘solar-powered’ sea slug, E. timida. Invertebr. Syst. 25, 477–489. ( 10.1071/IS11026) [DOI] [Google Scholar]
- 22.Mujer CV, Andrews DL, Manhart JR, Pierce SK, Rumpho ME. 1996. Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc. Natl Acad. Sci. USA 93, 12 333–12 338. ( 10.1073/pnas.93.22.12333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletreau KN, Weber APM, Weber KL, Rumpho ME. 2014. Lipid accumulation in the establishment of kleptoplasty in Elysia chlorotica. PLoS ONE 9, e97477 ( 10.1371/journal.pone.0097477.g012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrhenius A, Grönvall F, Scholze M, Backhaus T, Blanck H. 2004. Predictability of the mixture toxicity of 12 similarly acting congeneric inhibitors of photosystem II in marine periphyton and epipsammon communities. Aquat. Toxicol. 68, 351–367. ( 10.1016/j.aquatox.2004.04.002) [DOI] [PubMed] [Google Scholar]
- 25.Goldberg AL. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. ( 10.1038/nature02263) [DOI] [PubMed] [Google Scholar]
- 26.Kirkin V, McEwan DG, Novak I, Dikic I. 2009. A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269. ( 10.1016/j.molcel.2009.04.026) [DOI] [PubMed] [Google Scholar]
- 27.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco M-T, Moscat J. 2008. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 283, 6783–6789. ( 10.1074/jbc.M709496200) [DOI] [PubMed] [Google Scholar]
- 28.Balaban RS, Nemoto S, Finkel T. 2005. Mitochondria, oxidants, and aging. Cell 120, 483–495. ( 10.1016/j.cell.2005.02.001) [DOI] [PubMed] [Google Scholar]
- 29.Wägele H, Ballesteros M, Avila C. 2006. Defensive glandular structures in opisthobranch molluscs: from histology to ecology. Oceanogr. Mar. Biol. 44, 197–276. ( 10.1201/9781420006391.ch5) [DOI] [Google Scholar]
- 30.Carter WO, Narayanan PK, Robinson JP. 1994. Intracellular hydrogen peroxide and superoxide anion detection in endothelial cells. J. Leukoc. Biol. 55, 253–258. [DOI] [PubMed] [Google Scholar]
- 31.Cueto M, D'Croz L, Maté JL, San-Martín A, Darias J. 2005. Elysiapyrones from Elysia diomedea. Do such metabolites evidence an enzymatically assisted electrocyclization cascade for the biosynthesis of their bicycle[4.2.0]octane core? Org. Lett. 7, 415–418. ( 10.1021/ol0477428) [DOI] [PubMed] [Google Scholar]
- 32.Färber A, Young AJ, Ruban AV, Horton P, Jahns P. 1997. Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants: (the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching). Plant Phys. 115, 1609–1618. ( 10.1104/pp.115.4.1609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wägele H, Johnsen G. 2001. Observations on the histology and photosynthetic performance of ‘solar-powered’ opisthobranchs (Mollusca, Gastropoda, Opisthobranchia) containing symbiotic chloroplasts or zooxanthellae. Org. Divers. Evol. 1, 193–210. ( 10.1078/1439-6092-00016) [DOI] [Google Scholar]
- 34.Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. ( 10.1038/nmeth.2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Core Development Team 2014. R: a language and environment for statistical computing. See http://www.R-project.org. [Google Scholar]
- 36.Shapiro SS, Wilk MB. 1965. An analysis of variance test for normality (complete samples). Biometrika 52, 591–611. ( 10.1093/biomet/52.3-4.591) [DOI] [Google Scholar]
- 37.Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1, 50–60. ( 10.1214/aoms/1177730491) [DOI] [Google Scholar]
- 38.FastQC 2014. A quality control tool for high throughput sequence data See http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 4, 323 ( 10.1186/1471-2105-12-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 ( 10.1186/gb-2009-10-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–402. ( 10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2010. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135. ( 10.1093/nar/gkr1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. ( 10.1038/nature11413) [DOI] [PubMed] [Google Scholar]
- 48.Simakov O, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531. ( 10.1038/nature11696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeckmann B, et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370. ( 10.1093/nar/gkg095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278, 631–637. ( 10.1126/science.278.5338.631) [DOI] [PubMed] [Google Scholar]
- 51.Koonin EV, et al. 2004. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7 ( 10.1186/gb-2004-5-2-r7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. 2008. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. ( 10.1093/nar/gkm796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powell S, et al. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 42, D231–D239. ( 10.1093/nar/gkt1253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. ( 10.1093/nar/28.1.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. ( 10.1093/nar/gkm321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 134 ( 10.1186/1471-2105-13-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data was deposited at the NCBI Single Read Archive (SRP047359) and the Transcriptome Shotgun Assembly Sequence Database (E. cornigera: GBRW00000000; E. timida: GBRM00000000).