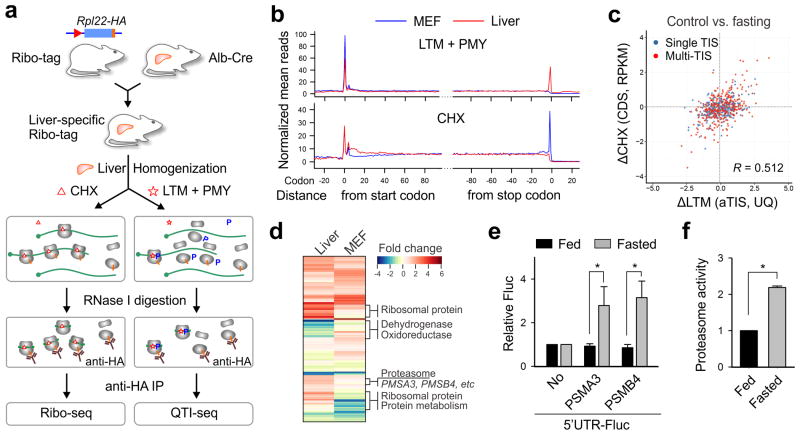
Figure 4. Liver-specific QTI-seq reveals translational reprogramming in response to fasting.
(a) Schematic of tissue-specific QTI-seq procedures using liver-specific RiboTag mice.
(b) Meta-gene analysis of LTM-associated ribosome density (top panel) or CHX-associated ribosome density (bottom panel) in MEF cells (blue line) and liver cells (red line). Normalized RPF reads are averaged across the entire transcriptome and aligned at the annotated start codons and stop codons.
(c) A scatter plot of fold changes in LTM-associated aTIS density and CHX-associated CDS ribosome occupancy in mouse liver cells with and without fasting.
(d) A heatmap of fold changes for gene groups with translational downregulation (green) or upregulation (red) in fasted liver (left) or starved MEF cells (right).
(e) Reporter assay using an in vitro translation system reprogrammed from mouse liver lysates with or without fasting. The relative translation efficiency of a synthesized Fluc mRNA containing 5′UTRs of PSMA3 or PSMB4 is shown in bar graph (means ± SEM; n = 3; * p < 0.01 student t-test).
(f) Mice of 8-12 week old were treated with or without overnight fasting. The chymotrypsin activity of liver homogenates was measured by Proteasome-Glo (means ± SEM; n = 3; * p < 0.01 student t-test).