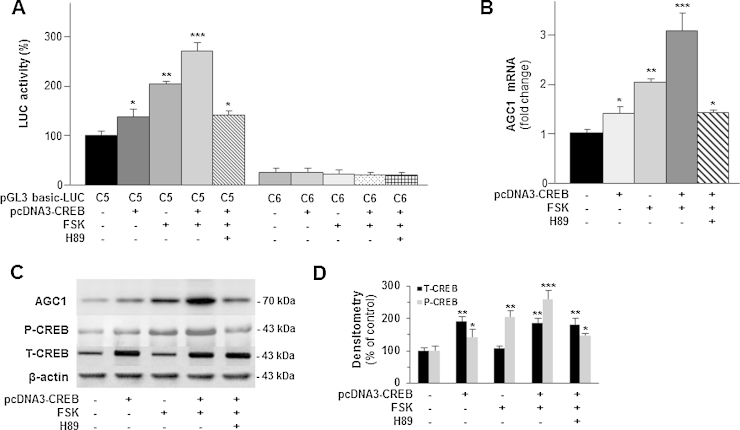
Fig. 3.
Functional characterization of CREB.
(A) SH-SY5Y transfected with pGL3-basic-Luc vector containing (C5) or not (C6) the CREB binding site. Where indicated a cotransfection with pcDNA3-CREB and/or a treatment with 10 μM forskolin for 3 h, and addition of 10 μM H89 1 h before forskolin were performed before luciferase assay. Data are expressed as means ± S.D. of five duplicate independent experiments.*P < 0.05, **P < 0.01, ***P < 0.001 versus control (one-way ANOVA). (B) Total RNA extracted from SH-SY5Y cells, which had been (where indicated) transfected with pcDNA3-CREB and/or treated with 10 μM forskolin for 1 h was used to quantify AGC1 mRNA by real-time PCR. Where indicated 10 μM H89 was added 1 h before forskolin. Data are expressed as means ± S.D. of five duplicate independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated control (one-way ANOVA). (C) AGC1, P-CREB, T-CREB and β-actin proteins of SH-SY5Y cells treated as above (B) were immunodetected with specific antibodies. P-CREB blot was reprobed for T-CREB quantification. A representative of four blots is shown. (D) Bands intensities of T-CREB and P-CREB were analyzed densitometrically using a Fluor-S MultiImager and Quantity One software (Bio-Rad) and corrected for β-actin. Data are expressed as means ± S.D. of four individual experiments. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated control.