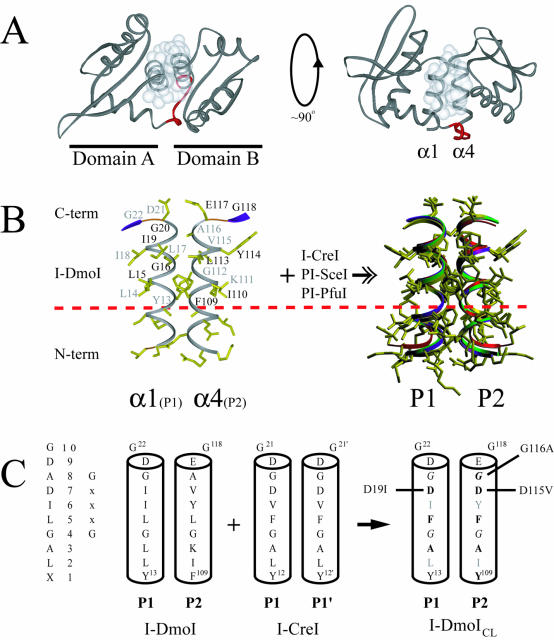
Figure 2.
Structure of I-DmoI and the LAGLIDADG motif. (A) Overall fold of I-DmoI. Left, top view down the two-helix bundle axis. Right, side view perpendicular to bundle axis. White spheres represent the van der Waals radii of the interfacial LAGLIDADG residues highlighting the majority of subdomain contacts between helices α1 and α4. Linker residues 98–105 are shown in red. (B) The α1/α4 helices (P1 and P2 regions) of I-DmoI with the XLAGLIDADG residues labeled. Black type, residues above the plane of the page; gray type, residues below the plane of the page. A dashed horizontal line demarcates the conserved sequence. At the right is the superposition of the LAGLIDADG helices of four known structures (I-DmoI, I-CreI, PI-SceI and PI-PfuI) demonstrating the extent of structural similarity in naturally occurring proteins. Although E-DreI, I-MsoI and I-AniI display a similar overlap, for clarity they were omitted from the superposition. (C) Creating I-DmoICL. The consensus sequence and generic numbering are shown on the left. The interfacial P1/P2 regions of I-DmoI were replaced by the P1/P1′ of I-CreI to generate I-DmoICL; bold type, residues changed in the LAGLIDADG of I-DmoI; gray type, residues facing the protein core and thus not changed to their I-CreI counterparts; italics, the GxxxG motif residues within the LAGLIDADG helices. Revertants are indicated to the left and right of the helices. See Figure 4 and the text for details.