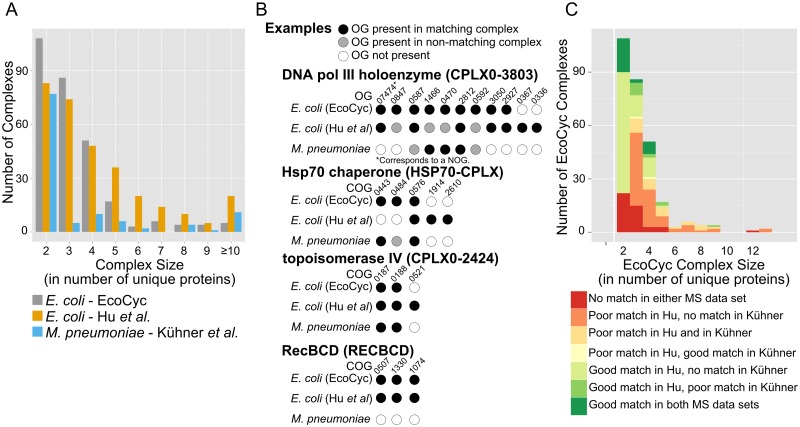
Fig 2. Protein complex data sets vary in composition.
(A) Count of complexes in two E. coli complex datasets ([5]; EcoCyc [13]) and one M. pneumoniae dataset [6], by size (in number of unique protein components). Multimers of single proteins (i.e., homodimers) are not included. (B) Examples of complex matching across data sets. Once mapped to an orthologous group (OG), the components of a complex are directly comparable to those in other complex sets yet perfect matches are rare. In some instances, an OG in one complex may not be present in its best matching complex but the OG may be present elsewhere in a different complex. In other cases, the matched complex may contain components (OGs) not seen in the query complex (as is the case with topoisomerase IV). (C) Summary of matching complex quality across data sets. EcoCyc complexes were used as the set of query complexes while the two experimental data sets were used as the search space. Here, a poor match requires just one matching component, while a good match requires at least half of the components in the query complex to be present in the matching complex. The number of complexes in each category is shown; complex size is as in part A.