Fig 10. Molecular dynamics simulations.
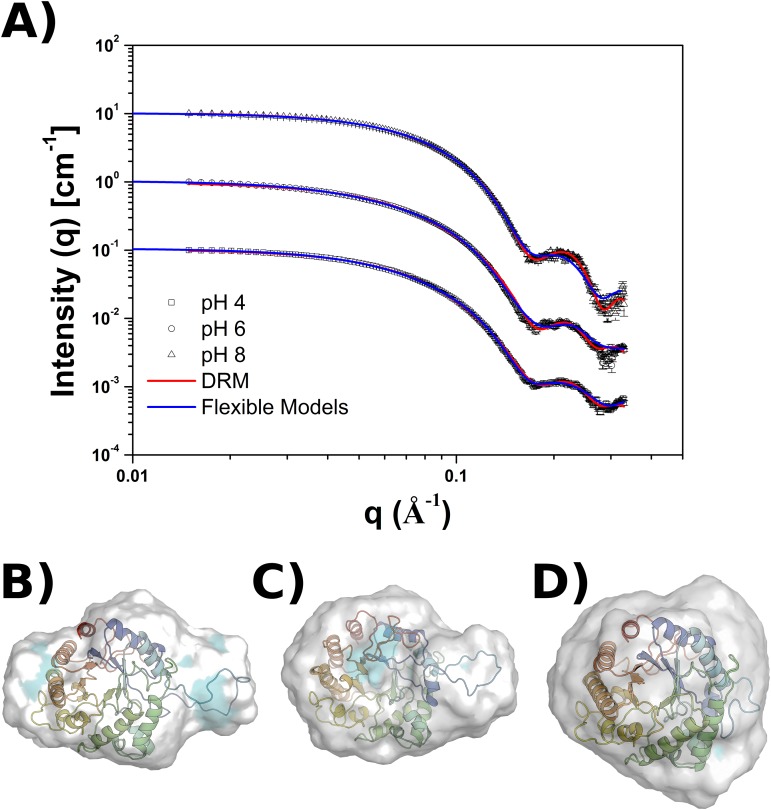
(A) Experimental and theoretical SAXS curves of the TpManGH5 at 20°C for the different pH scenarios. The theoretical curves were obtained from independent simulations and calculated using CRYSOL package. It is showed only the conformation with the lowest χ2 for each pH selected. The theoretical curves for flexible models (solid blue lines) and DRM (solid red lines) are overlapped in the experimental curves to the pH equals 8 (triangles), 6 (circles) and 4 (squares). The cartoon representations of the conformation with the best agreement with the experimental data are also presented. They show the models superimposed into the ab initio DRM low-resolution envelopes calculated using GASBOR to the pH 4 (B), 6 (C) and 8 (D). These Figures illustrate the variation in the elongation of the loop (residues Y88 to A105), indicating the region where conformational changes occurs and its flexibility for the different pHs. These results, combined with biochemical evaluation, suggest that the dynamics of the loop associated to the protonation state may define a balance for the optimal enzymatic activity. It also suggested that fluctuations in the loop may play an important role for the hydrolysis. Even, it is possible verify that the remaining part of the protein, i.e. the core of the protein, is not affected by the conformational changes of the loop. The blue spots in the low-resolution molecular envelopes indicate the shell hydration and support the hypothesis of the mechanical movement of the loop facilitate access to the water molecule, necessary during the hydrolysis.