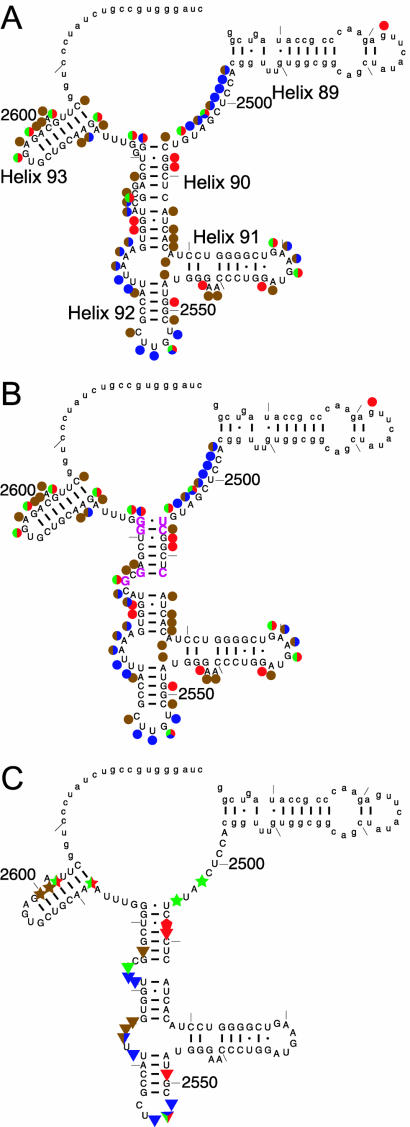
Figure 1.
(A) Cleavage and modification pattern of the 172mer by RNase T1 (green), RNase T2 (blue), kethoxal (red) and DMS (brown) shown on the phylogenetic secondary structure. (B) The cleavage/modification pattern shown on our proposed secondary structure. Rearranged residues are shown in magenta. (C) Footprinting of DbpA on the 172mer: apo-DbpA protection (triangles), AMPPNP-dependent DbpA protection (stars) and hypersensitivity (pentagons). Lower case denotes regions for which no data were collected.