Figure 2.
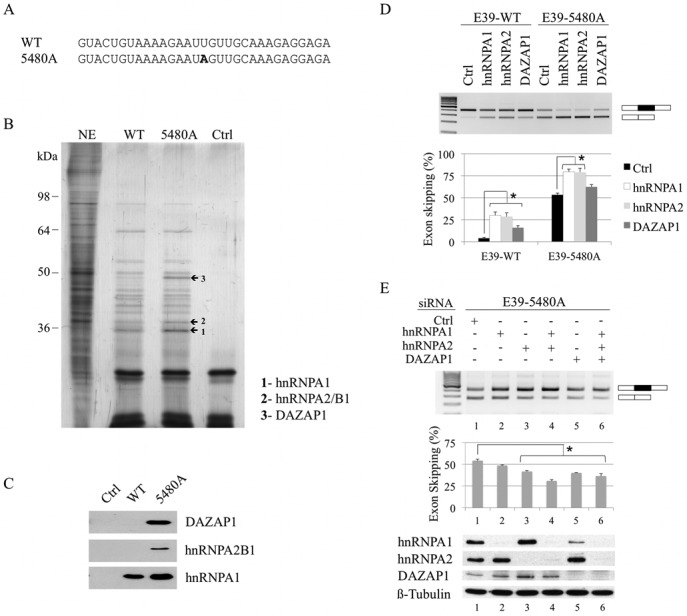
The splicing factors hnRNPA1, hnRNPA2B1 and DAZAP1 are preferentially recruited to the 5480A-mutated probe and promote exon 39 skipping. (A) Sequences of the WT and the mutated (5480A) RNA probes used for the RNA pull down experiments. The position of the 5480A mutation is in bold. (B) Study of hnRNPA1, hnRNPA2B1, and DAZAP1 binding by RNAse-assisted RNA pull down assay. The RNA probes were bound to adipic acid dihydrazide agarose beads and incubated with HeLa cell nuclear extract (NE). Proteins specifically bound to the RNA probes were eluted from beads by RNase treatment, separated on 10% SDS-PAGE gels and silver-stained. The proteins from bands 1 to 3 (arrows) were identified by mass spectrometry as hnRNPA1 (1), hnRNPA2B1 (2) and DAZAP1 (3). NE, nuclear extract; Ctrl, experiment performed without RNA probe. (C) Western blotting analysis of standard RNA pull down assays. The RNA probes bound to the adipic acid dihydrazide agarose beads were incubated with HeLa cell nuclear extract. After washing, pulled-down proteins were specifically detected by western blotting using antibodies directed against DAZAP1, hnRNPA2B1 and hnRNPA1. Ctrl, experiment performed without RNA probes. The experiment was repeated twice independently. (D) Effect of hnRNPA1, hnRNPA2 and DAZAP1 overexpression on exon 39 splicing. C25cl48 myoblasts were co-transfected with the E39-WT or E39-5480A minigene and hnRNPA1, hnRNPA2 and DAZAP1 expressing plasmids, or empty pcDNA3.1+ vector (control, Ctrl). Upper panel: RT-PCR analysis of exon 39 splicing pattern. The resulting products are indicated at the right side of the gel with alternatively spliced exon 39 (black box) and flanking vector exons (white boxes). Lower panel: quantification of exon 39 skipping percentage (mean ± SEM of four independent transfections). The result of the Wilcoxon rank-sum test is given (*P < 0.05) (E) Effect of hnRNPA1, hnRNPA2 and DAZAP1 depletion on mutant exon 39 splicing. After transfection with the siRNAs targeting hnRNPA1, hnRNPA2 or DAZAP1, C25cl48 myoblasts were transfected with the E39-5480A minigene. A non-targeting siRNA was used as negative control (Ctrl). Upper and middle panels: RT-PCR analysis, exon skipping quantification, and statistical analysis were as in Figure 2D. Lower panel: siRNA efficiency assessed by western blotting.
