Figure 3.
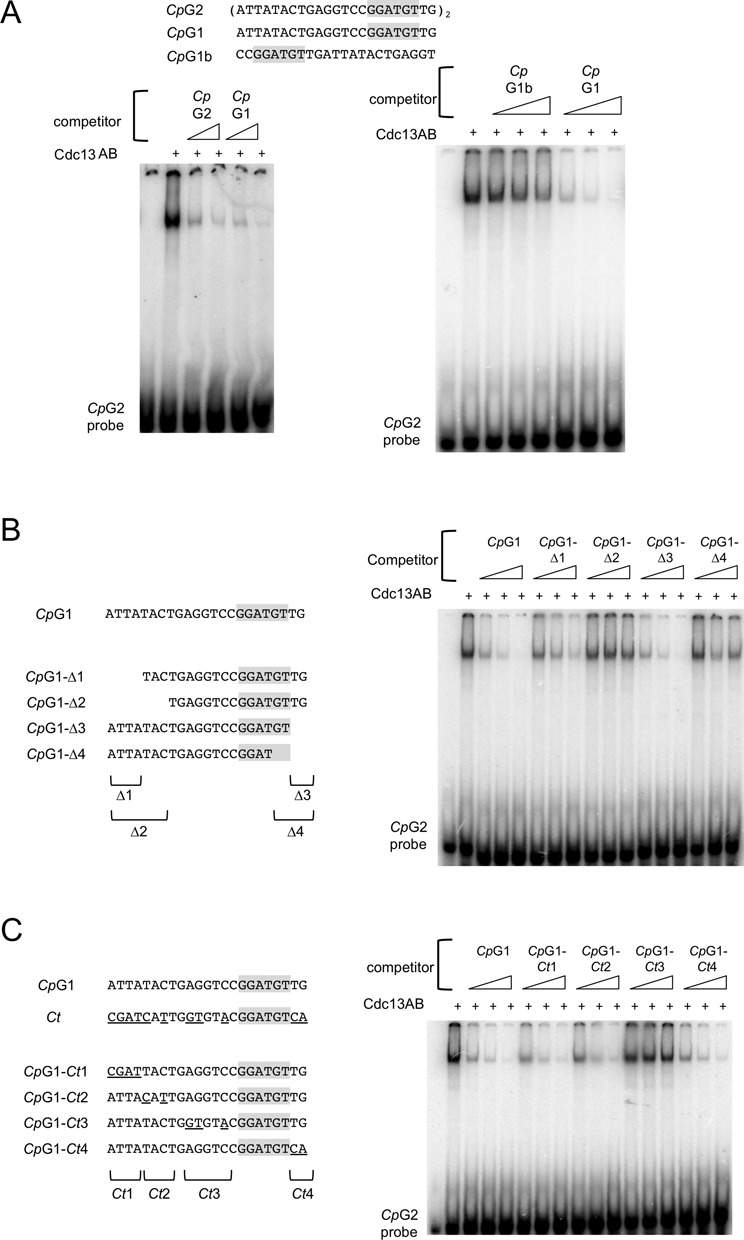
The sequence recognition property of the CpCdc13AB complex. (A) Left: Interaction between the CpG2 probe (7.5 nM) and the CpCdc13AB complex (10 nM) was assessed in gel mobility shift assays. Unlabeled CpG2 and CpG1 oligos (at 250 and 750 nM) were added to the assays as competitors. Right: Interaction between the CpG2 probe (7.5 nM) and the CpCdc13AB complex (10 nM) was assessed in gel mobility shift assays. Unlabeled CpG1b and CpG1 (at 23, 75 and 225 nM) were added to the assays as competitors. (B) Left: The truncation oligos used as competitors in the DNA-binding assays are illustrated. Right: Interaction between the CpG2 probe (7.5 nM) and the CpCdc13AB complex (10 nM) was assessed in gel mobility shift assays. Unlabeled CpG1-Δ1, Δ2, Δ3 and Δ4 oligos (at 23, 75 and 225 nM) were added to the assays as competitors. (C) Left: The hybrid Cp/Ct telomere oligos used as competitors in the DNA-binding assays are illustrated. Right: Interaction between the CpG2 probe (7.5 nM) and the CpCdc13AB complex (10 nM) was assessed in gel mobility shift assays. Unlabeled CpG1-Ct1, Ct2, Ct3 and Ct4 oligos (at 23, 75 and 225 nM) were added to the assays as competitors.
