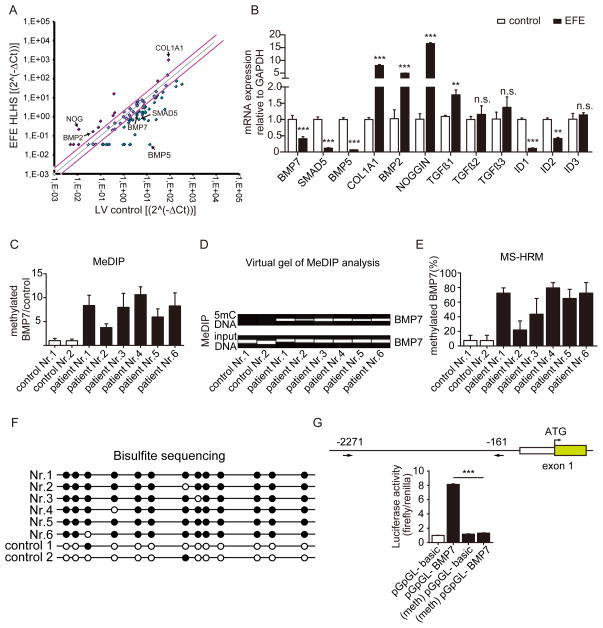
Figure 2. BMP7 promoter is hypermethylated in EFE.
(A) TGF-β/BMP qPCR Array compared EFE with healthy control heart tissues. The black arrow highlights the candidate genes which were altered in the EFE sample. (B) Quantitative real-time PCR analysis to validate the candidate genes selected from the TGF-β/BMP qPCR Array. (C, D) Methylated DNA immunoprecipitation (MeDIP) to assess BMP7 methylation in healthy and EFE hearts. (C) Real-time PCR analysis quantitatively showed the methylation levels in control and EFE samples and (D) virtual gel images of BMP7 PCR products (upper panel) of immunoprecipitated 5mC DNA and the lower panel shows PCR products of input DNA as loading controls. (E) BMP7 promoter methylation was determined by methylation-sensitive high resolution melting (MS-HRM) analysis. (F) BMP7 promoter methylation status of the individual CpG sites in the EFE samples and in healthy hearts by bisulfite sequencing. Open and filled circles indicate unmethylated and methylated status, respectively. (G) Luciferase assay assessed promoter activity of methylated human BMP7 promoter. Around 2.1kb (from 2271 to 161 upstream of transcriptional starting site) human BMP7 promoter fragment was cloned into a CpG- free luciferase vector (upper panel). The luciferase activity from the methylated BMP7 promoter construct was significantly reduced as compared to unmethylated control construct. Data are shown as the mean ± SD of three experiments. n.s. no significance, **, P<0.01, ***, P<0.001.